3IYV
 
 | | Clathrin D6 coat as full-length Triskelions | | Descriptor: | Clathrin heavy chain, Clathrin light chain A | | Authors: | Johnson, G.T, Fotin, A, Cheng, Y, Sliz, P, Grigorieff, N, Harrison, S.C, Kirchhausen, T, Walz, T. | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular model for a complete clathrin lattice from electron cryomicroscopy.
Nature, 432, 2004
|
|
1XI4
 
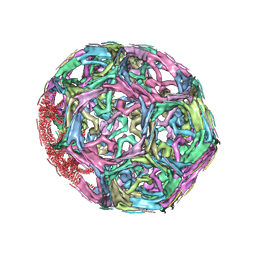 | | Clathrin D6 Coat | | Descriptor: | Clathrin heavy chain, Clathrin light chain A | | Authors: | Fotin, A, Cheng, Y, Sliz, P, Grigorieff, N, Harrison, S.C, Kirchhausen, T, Walz, T. | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular model for a complete clathrin lattice from electron cryomicroscopy
Nature, 432, 2004
|
|
1XI5
 
 | | Clathrin D6 coat with auxilin J-domain | | Descriptor: | Auxilin J-domain, Clathrin heavy chain | | Authors: | Fotin, A, Cheng, Y, Grigorieff, N, Walz, T, Harrison, S.C, Kirchhausen, T. | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating
Nature, 432, 2004
|
|
1I31
 
 | | MU2 ADAPTIN SUBUNIT (AP50) OF AP2 CLATHRIN ADAPTOR, COMPLEXED WITH EGFR INTERNALIZATION PEPTIDE FYRALM AT 2.5 A RESOLUTION | | Descriptor: | CLATHRIN COAT ASSEMBLY PROTEIN AP50, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Modis, Y, Boll, W, Rapoport, I, Kirchhausen, T. | | Deposit date: | 2001-02-12 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MU2 ADAPTIN SUBUNIT (AP50) OF AP2 CLATHRIN ADAPTOR, COMPLEXED WITH EGFR INTERNALIZATION PEPTIDE FYRALM AT 2.5 A RESOLUTION
To be Published
|
|
3N0A
 
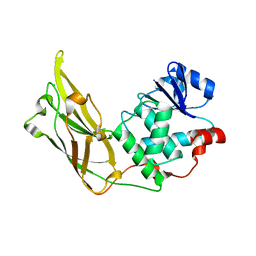 | | Crystal structure of auxilin (40-400) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein phosphatase auxilin | | Authors: | Harrison, S.C, Guan, R, Dai, H, Kirchhausen, T. | | Deposit date: | 2010-05-13 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the PTEN-like Region of Auxilin, a Detector of Clathrin-Coated Vesicle Budding.
Structure, 18, 2010
|
|
1BPO
 
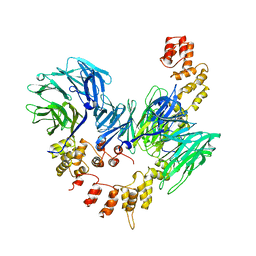 | | CLATHRIN HEAVY-CHAIN TERMINAL DOMAIN AND LINKER | | Descriptor: | PROTEIN (CLATHRIN) | | Authors: | Harr, E.T, Musacchio, A, Harrison, S.C, Kirchhausen, T. | | Deposit date: | 1998-08-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker.
Cell(Cambridge,Mass.), 95, 1998
|
|
1C9L
 
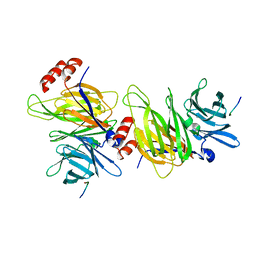 | |
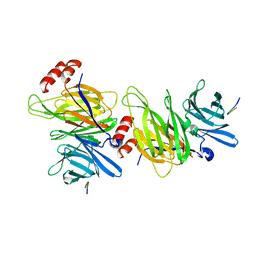
1C9I
 
 | |
1W63
 
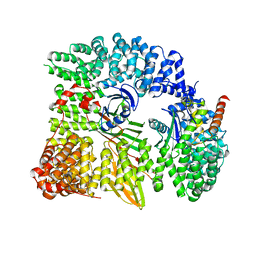 | | AP1 clathrin adaptor core | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 1 BETA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 SIGMA 1A SUBUNIT, ... | | Authors: | Heldwein, E, Macia, E, Wang, J, Yin, H.L, Kirchhausen, T, Harrison, S.C. | | Deposit date: | 2004-08-12 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of the Clathrin Adaptor Protein 1 Core
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7UPX
 
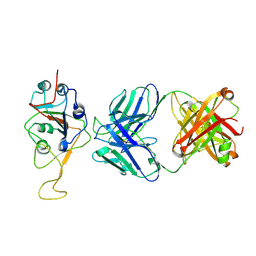 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SP1-77 Fab heavy chain, SP1-77 Fab light chain, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPY
 
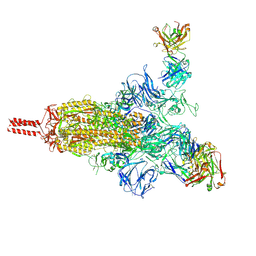 | | An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPW
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
3ML6
 
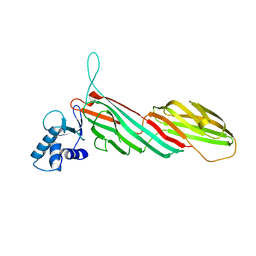 | | a complex between Dishevelled2 and clathrin adaptor AP-2 | | Descriptor: | Chimeric complex between protein Dishevelled2 homolog dvl-2 and clathrin adaptor AP-2 complex subunit mu | | Authors: | Yu, A, Xing, Y, Harrison, S.C, Kirchhausen, T.L. | | Deposit date: | 2010-04-16 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of the interaction between Dishevelled2 and clathrin AP-2 adaptor, a critical step in noncanonical Wnt signaling.
Structure, 18, 2010
|
|
8CZI
 
 | |
8FA2
 
 | |
8FA1
 
 | |
7TIK
 
 | |
2TBV
 
 | |
