5GKV
 
 | | Crystal Structure of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus | | Descriptor: | Esterase A | | Authors: | Ngo, T.D, Ryu, B.H, Kim, B.Y, Yoo, W.K, Lee, E.J, Lee, S.J, Kim, T.D, Kim, K.K. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Biochemical and Structural Analysis of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus
To Be Published
|
|
5HOE
 
 | | Crystal structrue of Est24, a carbohydrate acetylesterase from Sinorhizobium meliloti | | Descriptor: | HEXAETHYLENE GLYCOL, Hydrolase, PHOSPHATE ION | | Authors: | Oh, C, Ryu, B.H, An, D.R, Nguyen, D.D, Yoo, W, Kim, T, Ngo, D.T, Kim, H.S, Park, J.S, Kim, K.K, Kim, T.D. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Biochemical Characterization of an Octameric Carbohydrate Acetylesterase from Sinorhizobium meliloti.
Febs Lett., 590, 2016
|
|
5ZWR
 
 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | (2S)-2-[3-(benzenecarbonyl)phenyl]propanoic acid, Est-Y29, GLYCEROL | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Kim, K.K. | | Deposit date: | 2018-05-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
5ZWQ
 
 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | Est-Y29, GLYCEROL, ethyl (2S)-2-[3-(benzenecarbonyl)phenyl]propanoate | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W, Kim, K.K. | | Deposit date: | 2018-05-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
5ZWV
 
 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | Est-Y29 | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W. | | Deposit date: | 2018-05-17 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
2OZ4
 
 | | Structural Plasticity in IgSF Domain 4 of ICAM-1 Mediates Cell Surface Dimerization | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB FRAGMENT LIGHT CHAIN, ... | | Authors: | Chen, X, Kim, T.D, Carman, C.V, Mi, L, Song, G, Springer, T.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity in Ig superfamily domain 4 of ICAM-1 mediates cell surface dimerization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4TX1
 
 | |
7C4X
 
 | |
4P6B
 
 | | Crystal structure of Est-Y29,a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-24 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P85
 
 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | DIETHYL PHOSPHONATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P87
 
 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5H3H
 
 | | Esterase (EaEST) from Exiguobacterium antarcticum | | Descriptor: | Abhydrolase domain-containing protein, ETHANEPEROXOIC ACID | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-10-24 | | Release date: | 2017-01-11 | | Last modified: | 2022-10-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Functional Characterization of an Esterase (EaEST) from Exiguobacterium antarcticum.
Plos One, 12, 2017
|
|
6IYM
 
 | |
6JZL
 
 | |
6AGQ
 
 | | Acetyl xylan esterase from Paenibacillus sp. R4 | | Descriptor: | ZINC ION, acetyl xylan esterase | | Authors: | Park, S, Lee, C.W, Lee, J.H. | | Deposit date: | 2018-08-13 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional characterization of a cold-active acetyl xylan esterase (PbAcE) from psychrophilic soil microbe Paenibacillus sp.
PLoS ONE, 13, 2018
|
|
1FK2
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH MYRISTIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, MYRISTIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK7
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH RICINOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN, RICINOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK3
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH PALMITOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK0
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH CAPRIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | DECANOIC ACID, FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-08 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK5
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH OLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, OLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK4
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH STEARIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, STEARIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK1
 
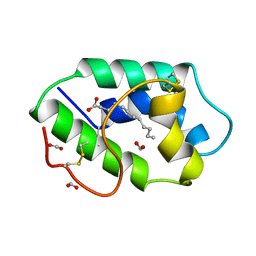 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH LAURIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, LAURIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK6
 
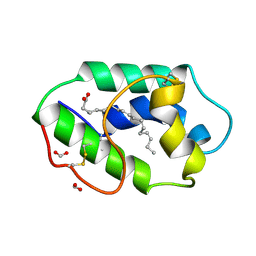 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH ALPHA-LINOLENIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | ALPHA-LINOLENIC ACID, FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
