3CZG
 
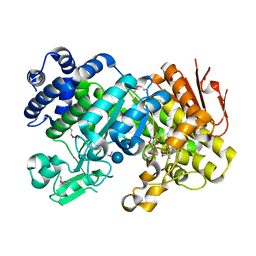 | | Crystal Structure Analysis of Sucrose hydrolase (SUH)-glucose complex | | Descriptor: | Sucrose hydrolase, alpha-D-glucopyranose | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
3CZE
 
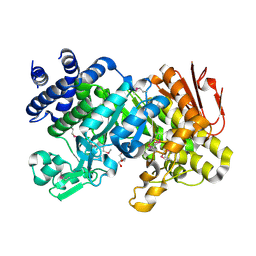 | | Crystal Structure Analysis of Sucrose hydrolase (SUH)- Tris complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Sucrose hydrolase | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
3CZL
 
 | |
3CZK
 
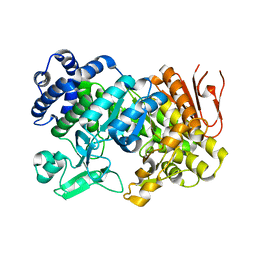 | | Crystal Structure Analysis of Sucrose hydrolase(SUH) E322Q-sucrose complex | | Descriptor: | Sucrose hydrolase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
5B6A
 
 | | Structure of Pyridoxal Kinasefrom Pseudomonas Aeruginosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kim, M.I, Hong, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and catalytic mechanism of pyridoxal kinase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 478, 2016
|
|
4FJS
 
 | | Crystal structure of ureidoglycolate dehydrogenase enzyme in apo form | | Descriptor: | Ureidoglycolate dehydrogenase | | Authors: | Kim, M.I, Shin, I, Lee, J, Rhee, S. | | Deposit date: | 2012-06-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
4FJU
 
 | |
4ZPX
 
 | |
3OUL
 
 | |
3OUM
 
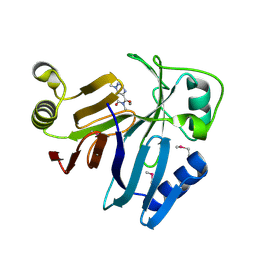 | | Crystal Structure of toxoflavin-degrading enzyme in complex with toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, MANGANESE (II) ION, toxoflavin-degrading enzyme | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2010-09-15 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of phytotoxin toxoflavin-degrading enzyme
Plos One, 6, 2011
|
|
4XRF
 
 | |
5IE9
 
 | |
4H8A
 
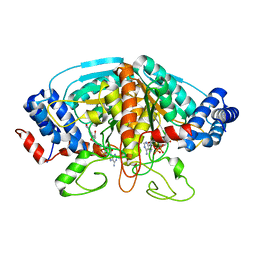 | | Crystal structure of ureidoglycolate dehydrogenase in binary complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ureidoglycolate dehydrogenase | | Authors: | Rhee, S, Shin, I, Kim, M. | | Deposit date: | 2012-09-22 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
7EYC
 
 | | Crystal structure of Tau and acetylated tau peptide antigen | | Descriptor: | ACETYLATED TAU PEPTIDE, antibody, Heavy chain, ... | | Authors: | Hong, M, Park, J. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Monoclonal antibody Y01 prevents tauopathy progression induced by lysine 280-acetylated tau in cell and mouse models.
J.Clin.Invest., 133, 2023
|
|
7BWL
 
 | | Structure of antibiotic sequester from Pseudomonas aerurinosa | | Descriptor: | UPF0312 protein PA0423, Ubiquinone-8 | | Authors: | Hong, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Pseudomonas aeruginosa PA0423 protein and its functional implication in antibiotic sequestration.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
5ZQH
 
 | | Crystal structure of Streptococcus transcriptional regulator | | Descriptor: | PadR family transcriptional regulator | | Authors: | Kim, M, Hong, M. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based functional analysis of a PadR transcription factor from Streptococcus pneumoniae and characteristic features in the PadR subfamily-2.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
4FFH
 
 | |
4FFG
 
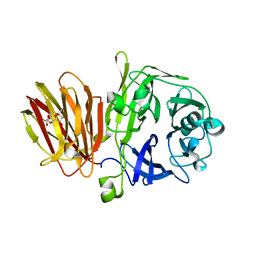 | | Crystal Structure of Levan Fructotransferase from Arthrobacter ureafaciens in complex with DFA-IV | | Descriptor: | (1R,4R,5S,6S,7R,10R,11S,12S)-1,7-bis(hydroxymethyl)-2,8,13,14-tetraoxatricyclo[8.2.1.1~4,7~]tetradecane-5,6,11,12-tetrol, Levan fructotransferase, beta-D-fructofuranose-(2-6)-beta-D-fructofuranose | | Authors: | Park, J, Rhee, S. | | Deposit date: | 2012-06-01 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional basis for substrate specificity and catalysis of levan fructotransferase.
J.Biol.Chem., 287, 2012
|
|
4FFF
 
 | |
4FFI
 
 | |
3E74
 
 | |
5H20
 
 | | X-ray structure of PadR-like Transcription factor from bacteroid fragilis | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHATE ION, Putative PadR-family transcriptional regulatory protein, ... | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2016-10-13 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional analysis of BF2549, a PadR-like transcription factor from Bacteroides fragilis.
Biochem. Biophys. Res. Commun., 483, 2017
|
|
3RZU
 
 | | The Crystal Structure of the Catalytic Domain of AMSH | | Descriptor: | STAM-binding protein, ZINC ION | | Authors: | Davies, C.W, Das, C. | | Deposit date: | 2011-05-12 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Thermodynamic Comparison of the Catalytic Domain of AMSH and AMSH-LP: Nearly Identical Fold but Different Stability.
J.Mol.Biol., 413, 2011
|
|
3RZV
 
 | |
5XEF
 
 | | Crystal structure of flagellar chaperone from bacteria | | Descriptor: | Flagellar protein fliS | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-27 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the flagellar chaperone FliS from Bacillus cereus and an invariant proline critical for FliS dimerization and flagellin recognition
Biochem. Biophys. Res. Commun., 487, 2017
|
|
