6L0Y
 
 | | Structure of dsRNA with G-U wobble base pairs | | Descriptor: | RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3') | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of dsRNA with G-U wobble base pairs
To Be Published
|
|
5ET3
 
 | | Crystal Structure of De novo Designed Fullerene organizing peptide | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, Fullerene Organizing Protein (C60Sol-COP-3) | | Authors: | Kim, K.-H, Kim, Y.H, Acharya, R, Kim, N.H, Paul, J, Grigoryan, G, DeGrado, W.F. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.671 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
3OIC
 
 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (apo form) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], SULFATE ION | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OID
 
 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (complex with NADP and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OIF
 
 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OIG
 
 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and INH) | | Descriptor: | (2E)-N-[(1,2-dimethyl-1H-indol-3-yl)methyl]-N-methyl-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3S2P
 
 | | Crystal structure of CDK2 with a 2-aminopyrimidine compound | | Descriptor: | (3S,4S)-1-{3-[2-amino-6-(propan-2-yl)pyrimidin-4-yl]-4-hydroxyphenyl}pyrrolidine-3,4-diol, Cyclin-dependent kinase 2 | | Authors: | Kim, K.-H, Lee, J, Jeong, S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel class of 2-aminopyrimidines as CDK1 and CDK2 inhibitors
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2OS1
 
 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | ACTINONIN, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Park, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
2OS3
 
 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | ACTINONIN, COBALT (II) ION, Peptide deformylase | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Parh, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
2OS0
 
 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Park, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
1IC2
 
 | | DECIPHERING THE DESIGN OF THE TROPOMYOSIN MOLECULE | | Descriptor: | TROPOMYOSIN ALPHA CHAIN, SKELETAL MUSCLE | | Authors: | Brown, J.H, Kim, K.-H, Jun, G, Greenfield, N.J, Dominguez, R, Volkmann, N, Hitchcock-DeGregori, S.E, Cohen, C. | | Deposit date: | 2001-03-29 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the design of the tropomyosin molecule
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7ENY
 
 | | Crystal structure of hydroxysteroid dehydrogenase from Escherichia coli | | Descriptor: | 7alpha-hydroxysteroid dehydrogenase | | Authors: | Kim, K.-H, Lee, C.W, Pardhe, D.P, Hwang, J, Do, H, Lee, Y.M, Lee, J.H, Oh, T.-J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of an apo 7 alpha-hydroxysteroid dehydrogenase reveals key structural changes induced by substrate and co-factor binding.
J.Steroid Biochem.Mol.Biol., 212, 2021
|
|
6KYV
 
 | | Crystal Structure of RIG-I and hairpin RNA with G-U wobble base pairs | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3'), ZINC ION | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Rna Biol., 17, 2020
|
|
2QKY
 
 | | complex structure of dipeptidyl peptidase IV and a oxadiazolyl ketone | | Descriptor: | 2-[(2-{(2S,4S)-2-[(R)-(5-tert-butyl-1,3,4-oxadiazol-2-yl)(hydroxy)methyl]-4-fluoropyrrolidin-1-yl}-2-oxoethyl)amino]-2-methylpropan-1-ol, Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) (Dipeptidyl peptidase 4 soluble form) (Dipeptidyl peptidase IV soluble form) | | Authors: | Kim, K.-H, Hong, S.Y, Koo, K.D, Lee, C.-S, Kim, G.T, Han, H.O. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis, SAR, and X-ray structure of novel potent DPPIV inhibitors: oxadiazolyl ketones.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1OH0
 
 | | CRYSTAL STRUCTURE OF KETOSTEROID ISOMERASE COMPLEXED WITH EQUILENIN | | Descriptor: | BETA-MERCAPTOETHANOL, EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Kim, K.-H, Cha, S.-S, Byun, M, Oh, B.-H. | | Deposit date: | 2003-05-21 | | Release date: | 2003-06-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Crystal Structures of Delta5-3-Ketosteroid Isomerase with and without a Reaction Intermediate Analogue
Biochemistry, 36, 1997
|
|
1NCO
 
 | | STRUCTURE OF THE ANTITUMOR PROTEIN-CHROMOPHORE COMPLEX NEOCARZINOSTATIN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, HOLO-NEOCARZINOSTATIN, NEOCARZINOSTATIN-CHROMOPHORE | | Authors: | Kim, K.-H, Kwon, B.-M, Myers, A.G, Rees, D.C. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of neocarzinostatin, an antitumor protein-chromophore complex.
Science, 262, 1993
|
|
4PQQ
 
 | | The crystal structure of discoidin domain from muskelin | | Descriptor: | Muskelin, PHOSPHATE ION, TETRAETHYLENE GLYCOL | | Authors: | Kim, K.-H, Hong, S.K, Kim, E.E. | | Deposit date: | 2014-03-04 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of mouse muskelin discoidin domain and biochemical characterization of its self-association.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3RBY
 
 | |
1H3Q
 
 | |
5HKN
 
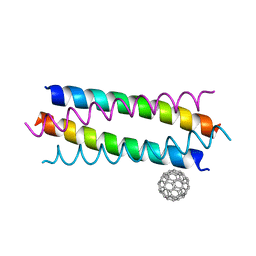 | | Crystal structure de novo designed fullerene organizing protein complex with fullerene | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | Authors: | Paul, J, Acharya, R, Kim, K.-H, Kim, Y.H, Kim, N.H, Grigoryan, G, DeGardo, W.F. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
5JI7
 
 | |
5JI9
 
 | |
5JIU
 
 | |
5JIA
 
 | |
5XNT
 
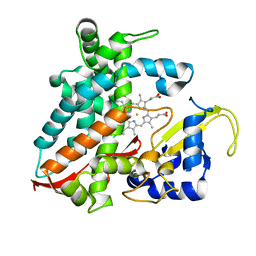 | | Structure of CYP106A2 from Bacillus sp. PAMC 23377 | | Descriptor: | Cytochrome P450 CYP106, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, C.W, Kim, K.-H, Bikash, D, Park, S.-H, Park, H, Oh, T.-J, Lee, J.H. | | Deposit date: | 2017-05-24 | | Release date: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Functional Characterization of a Cytochrome P450 (BaCYP106A2) fromBacillussp. PAMC 23377.
J. Microbiol. Biotechnol., 27, 2017
|
|
