1OAV
 
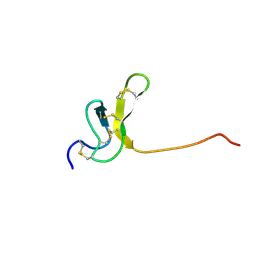 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
1OAW
 
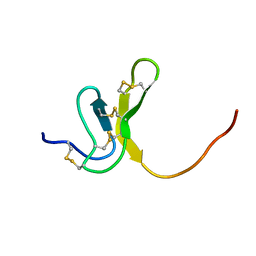 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
2B8I
 
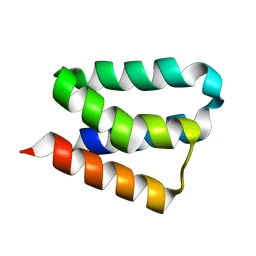 | | Crystal Structure and Functional Studies Reveal that PAS Factor from Vibrio vulnificus is a Novel Member of the Saposin-Fold Family | | Descriptor: | PAS factor | | Authors: | Lee, J.H, Yang, S.T, Rho, S.H, Im, Y.J, Kim, S.Y, Kim, Y.R, Kim, M.K, Kang, G.B, Kim, J.I, Rhee, J.H, Eom, S.H. | | Deposit date: | 2005-10-07 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and functional studies reveal that PAS factor from Vibrio vulnificus is a novel member of the saposin-fold family
J.Mol.Biol., 355, 2006
|
|
2MZ4
 
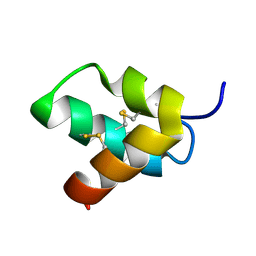 | | Solution Structure of mu-SLPTX-Ssm6a | | Descriptor: | Mu-scoloptoxin-Ssm6a | | Authors: | Kim, J.H, Kim, J.I. | | Deposit date: | 2015-02-06 | | Release date: | 2016-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of mu-SLPTX-Ssm6a, a Gating Modifier of human Nav1.7 channels
To be Published
|
|
2N9Z
 
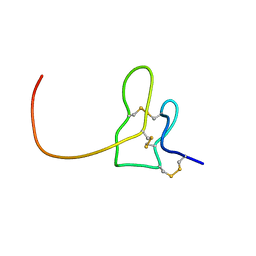 | | Solution structure of K1 lobe of double-knot toxin | | Descriptor: | Tau-theraphotoxin-Hs1a | | Authors: | Bae, C, Anselmi, C, Kalia, J, Jara-Oseguera, A, Schwieters, C.D, Krepkiy, D, Lee, C.W, Kim, E.H, Kim, J.I, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin
Elife, 5, 2016
|
|
2NAJ
 
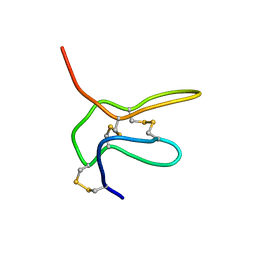 | | Solution structure of K2 lobe of double-knot toxin | | Descriptor: | Tau-theraphotoxin-Hs1a | | Authors: | Bae, C, Anselmi, C, Kalia, J, Jara-Oseguera, A, Schwieters, C.D, Krepkiy, D, Lee, C.W, Kim, E.H, Kim, J.I, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin
Elife, 5, 2016
|
|
2WH9
 
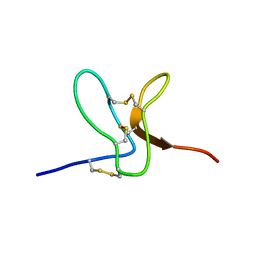 | | Solution structure of GxTX-1E | | Descriptor: | GUANGXITOXIN-1EGXTX-1E | | Authors: | Lee, S.K, Jung, H.H, Lee, J.Y, Lee, C.W, Kim, J.I. | | Deposit date: | 2009-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Gxtx-1E, a High Affinity Tarantula Toxin Interacting with Voltage Sensors in Kv2.1 Potassium Channels.
Biochemistry, 49, 2010
|
|
1S6X
 
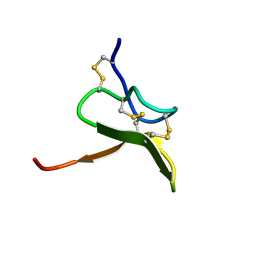 | | Solution structure of VSTx | | Descriptor: | KvAP CHANNEL | | Authors: | Jung, H.J, Eu, Y.J, Kim, J.I. | | Deposit date: | 2004-01-28 | | Release date: | 2005-03-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure and lipid membrane partitioning of VSTx1, an inhibitor of the KvAP potassium channel.
Biochemistry, 44, 2005
|
|
2NDB
 
 | |
1IE6
 
 | | SOLUTION STRUCTURE OF IMPERATOXIN A | | Descriptor: | IMPERATOXIN A | | Authors: | Lee, C.W, Takeuchi, K, Takahashi, H, Sato, K, Shimada, I, Kim, D.H, Kim, J.I. | | Deposit date: | 2001-04-07 | | Release date: | 2003-06-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A.
Biochem.J., 377, 2004
|
|
4M4D
 
 | | Crystal structure of lipopolysaccharide binding protein | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipopolysaccharide-binding protein | | Authors: | Eckert, J.K, Kim, Y.J, Kim, J.I, Gurtler, K, Oh, D.Y, Ploeg, A.H, Pickkers, P, Lundvall, L, Hamann, L, Giamarellos-Bourboulis, E, Kubarenko, A.V, Weber, A.N, Kabesch, M, Kumpf, O, An, H.J, Lee, J.O, Schumann, R.R. | | Deposit date: | 2013-08-07 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | The crystal structure of lipopolysaccharide binding protein reveals the location of a frequent mutation that impairs innate immunity.
Immunity, 39, 2013
|
|
1LA4
 
 | | Solution Structure of SGTx1 | | Descriptor: | SGTx1 | | Authors: | Lee, C.W, Roh, S.H, Kim, S, Endoh, H, Kodera, Y, Maeda, T, Swartz, K.J, Kim, J.I. | | Deposit date: | 2002-03-28 | | Release date: | 2003-11-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Characterization of SGTx1, a Modifier of Kv2.1 Channel Gating
Biochemistry, 43, 2004
|
|
1D1H
 
 | | SOLUTION STRUCTURE OF HANATOXIN 1 | | Descriptor: | HANATOXIN TYPE 1 | | Authors: | Takahashi, H, Kim, J.I, Sato, K, Swartz, K.J, Shimada, I. | | Deposit date: | 1999-09-16 | | Release date: | 2000-09-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hanatoxin1, a gating modifier of voltage-dependent K(+) channels: common surface features of gating modifier toxins.
J.Mol.Biol., 297, 2000
|
|
1T1T
 
 | | Solution Structure of Kurtoxin | | Descriptor: | Kurtoxin | | Authors: | Lee, C.W, Min, H.J, Cho, E.M, Kohno, T, Eu, Y.J, Kim, J.I. | | Deposit date: | 2004-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kurtoxin
To be Published
|
|
1X8D
 
 | | Crystal structure of E. coli YiiL protein containing L-rhamnose | | Descriptor: | Hypothetical protein yiiL, L-RHAMNOSE | | Authors: | Ryu, K.S, Kim, J.I, Cho, S.J, Park, D, Park, C, Lee, J.O, Choi, B.S. | | Deposit date: | 2004-08-18 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Monosaccharide Specificity of Escherichia coli Rhamnose Mutarotase
J.Mol.Biol., 349, 2005
|
|
2JRV
 
 | | The third dimensional structure of mab198-bound pep.1 for autoimmune myasthenia gravis | | Descriptor: | PEPTIDE PEP.1 | | Authors: | Jung, H.H, Yi, H.J, Lee, S.K, Lee, J.Y, Jung, H.J, Yang, S.T, Eu, Y.-J, Im, S.-H, Kim, J.I. | | Deposit date: | 2007-06-29 | | Release date: | 2008-07-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of immunotherapeutic peptides for autoimmune Myasthenia gravis
Biochemistry, 46, 2007
|
|
2JRW
 
 | | Solution structure of Cyclic extended Pep1(Cyc.ext.Pep.1) for autoimmune myasthenia gravis | | Descriptor: | Cyclic extended Pep.1 | | Authors: | Jung, H.H, Yi, H.J, Lee, S.K, Lee, J.Y, Jung, H.J, Yang, S.T, Eu, Y.-J, Im, S.-H, Kim, J.I. | | Deposit date: | 2007-06-29 | | Release date: | 2008-07-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of immunotherapeutic peptides for autoimmune Myasthenia gravis
Biochemistry, 46, 2007
|
|
2NDD
 
 | |
1LU8
 
 | |
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
1R02
 
 | |
6HHO
 
 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
5GWG
 
 | | Solution structure of rattusin | | Descriptor: | Defensin alpha-related sequence 1 | | Authors: | Lee, C.W, Min, H.J. | | Deposit date: | 2016-09-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rattusin structure reveals a novel defensin scaffold formed by intermolecular disulfide exchanges
Sci Rep, 7, 2017
|
|
1OMG
 
 | | NMR STUDY OF OMEGA-CONOTOXIN MVIIA | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Kohno, T, Kim, J.-I, Kobayashi, K, Kodera, Y, Maeda, T, Sato, K. | | Deposit date: | 1995-04-26 | | Release date: | 1996-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the calcium channel blocker omega-conotoxin MVIIA.
Biochemistry, 34, 1995
|
|
1DLF
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | Descriptor: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
