3OLM
 
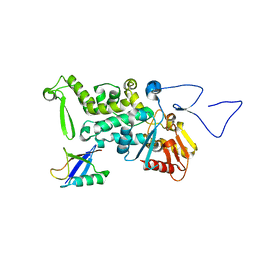 | | Structure and Function of a Ubiquitin Binding Site within the Catalytic Domain of a HECT Ubiquitin Ligase | | Descriptor: | E3 ubiquitin-protein ligase RSP5, Ubiquitin | | Authors: | Kim, H.C, Steffen, A, Chen, J, Huibregtse, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structure and function of a HECT domain ubiquitin-binding site.
Embo Rep., 12, 2011
|
|
4M6R
 
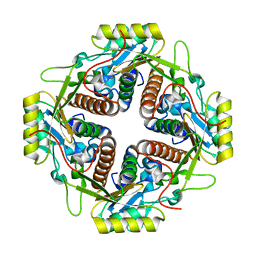 | | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme | | Descriptor: | Methylthioribulose-1-phosphate dehydratase, ZINC ION | | Authors: | Kang, W, Hong, S.H, Lee, H.M, Kim, N.Y, Lim, Y.C, Le, L.T.M, Lim, B, Kim, H.C, Kim, T.Y, Ashida, H, Yokota, A, Hah, S.S, Chun, K.H, Jung, Y.K, Yang, J.K. | | Deposit date: | 2013-08-10 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7QI8
 
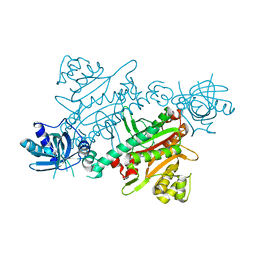 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE AND INHIBITOR | | Descriptor: | 2-azanyl-6-[(1~{S},7~{S})-2,2-bis(fluoranyl)-7-oxidanyl-cycloheptyl]-4-methoxy-7~{H}-pyrrolo[3,4-d]pyrimidin-5-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7QHN
 
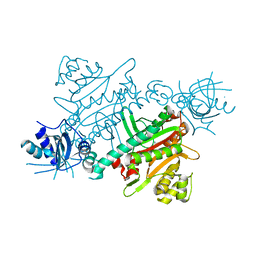 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE and an inhibitor | | Descriptor: | 6-azanyl-2-cyclohexyl-4-fluoranyl-1~{H}-pyrrolo[3,4-c]pyridin-3-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7QH8
 
 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE | | Descriptor: | GLYCEROL, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
5DMV
 
 | | Polo-box domain of Mouse Polo-like kinase 1 complexed with Emi2 (146-177) | | Descriptor: | F-box only protein 43, NICKEL (II) ION, Serine/threonine-protein kinase PLK1 | | Authors: | Namgoong, S. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis for recognition of Emi2 by Polo-like kinase 1 and development of peptidomimetics blocking oocyte maturation and fertilization.
Sci Rep, 5, 2015
|
|
5DNJ
 
 | | Mouse Polo-box domain and Peptide analog 702 | | Descriptor: | Serine/threonine-protein kinase PLK1, peptide 707-56A-SER-TPO-NH2 | | Authors: | Namgoong, S, Han, Y.H. | | Deposit date: | 2015-09-10 | | Release date: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of Emi2 by Polo-like kinase 1 and development of peptidomimetics blocking oocyte maturation and fertilization.
Sci Rep, 5, 2015
|
|
5DMS
 
 | | Mouse Polo-box domain and Emi2 (169-177) | | Descriptor: | F-box only protein 43, Serine/threonine-protein kinase PLK1 | | Authors: | Namgoong, S. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.89999 Å) | | Cite: | Structural basis for recognition of Emi2 by Polo-like kinase 1 and development of peptidomimetics blocking oocyte maturation and fertilization.
Sci Rep, 5, 2015
|
|
1L9M
 
 | |
1L9N
 
 | |
1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2G
 
 | | Sir2 homologue-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2J
 
 | | Sir2 homologue H80N mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2H
 
 | | Sir2 homologue S24A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2N
 
 | |
1NUG
 
 | |
1NUF
 
 | |
1NUD
 
 | |
