7X89
 
 | | Tid1 | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Jang, J, Lee, S.H, Kang, D.H, Sim, D.W, Jo, K.S, Ryu, H, Kim, E.H, Ryu, K.S, Lee, J.H, Kim, J.H, Won, H.S. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the J-domain and GF-motif of the mitochondrial Hsp40, Tid1
To Be Published
|
|
2N9Z
 
 | | Solution structure of K1 lobe of double-knot toxin | | Descriptor: | Tau-theraphotoxin-Hs1a | | Authors: | Bae, C, Anselmi, C, Kalia, J, Jara-Oseguera, A, Schwieters, C.D, Krepkiy, D, Lee, C.W, Kim, E.H, Kim, J.I, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin
Elife, 5, 2016
|
|
2NAJ
 
 | | Solution structure of K2 lobe of double-knot toxin | | Descriptor: | Tau-theraphotoxin-Hs1a | | Authors: | Bae, C, Anselmi, C, Kalia, J, Jara-Oseguera, A, Schwieters, C.D, Krepkiy, D, Lee, C.W, Kim, E.H, Kim, J.I, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin
Elife, 5, 2016
|
|
5J4F
 
 | | Crystal structure of the N-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.-W, Lee, W.-C, Kim, H.Y, Kim, J.-H, Won, H.-S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
5J4G
 
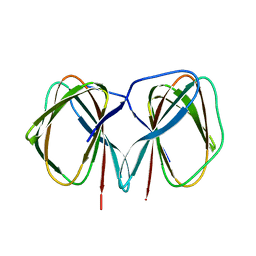 | | Crystal structure of the C-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.W, Lee, W.C, Kim, H.Y, Kim, J.H, Won, H.S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
5XV9
 
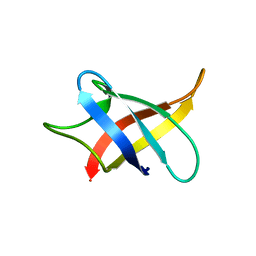 | |
7VZM
 
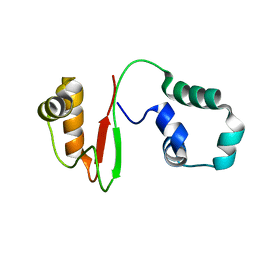 | | Anti-CRISPR AcrIE4-F7 | | Descriptor: | AcrIE4-F7 | | Authors: | Hong, S.H, Lee, G, Bae, E, Suh, J.Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-02-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The structure of AcrIE4-F7 reveals a common strategy for dual CRISPR inhibition by targeting PAM recognition sites.
Nucleic Acids Res., 50, 2022
|
|
2MJ6
 
 | |
2LXJ
 
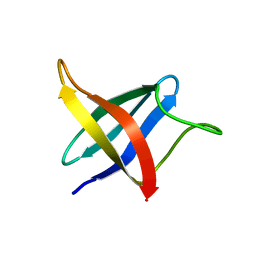 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp with dT7 | | Descriptor: | Cold shock-like protein CspLA | | Authors: | Lee, J, Jeong, K, Kim, Y. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
2LXK
 
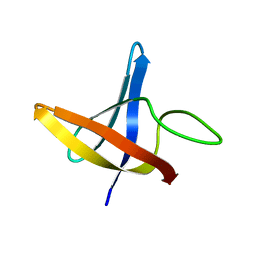 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp | | Descriptor: | Cold shock-like protein CspLA | | Authors: | Lee, J, Jeong, K, Kim, Y. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
2MNU
 
 | |
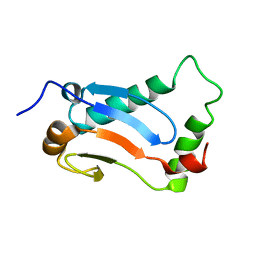
2MUK
 
 | |
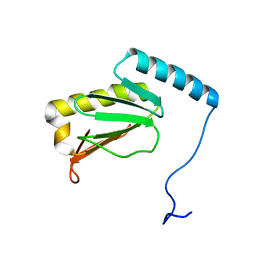
6LKF
 
 | |
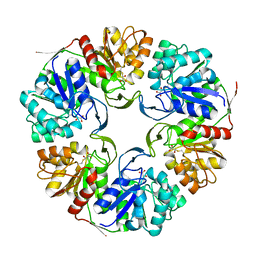
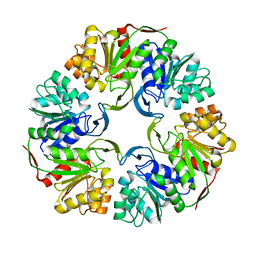
4OGG
 
 | |
4OFW
 
 | |
4OGF
 
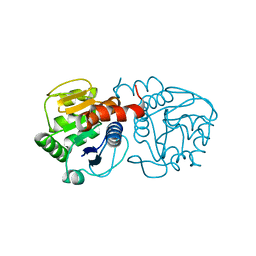 | |
6M3N
 
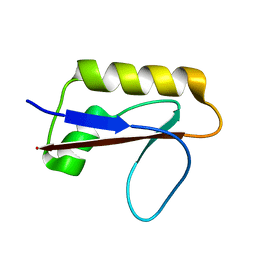 | | Solution structure of anti-CRISPR AcrIF7 | | Descriptor: | anti-CRIPSR AcrIF7 | | Authors: | Kim, I, An, S.Y, Koo, J, Bae, E, Suh, J.Y. | | Deposit date: | 2020-03-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into the CRISPR inhibition of AcrIF7.
Nucleic Acids Res., 48, 2020
|
|
3OZQ
 
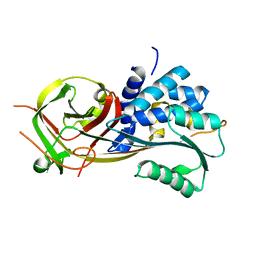 | |
7KP6
 
 | | Structure of Ack1 kinase in complex with a selective inhibitor | | Descriptor: | 5-chloro-N~2~-[4-(4-methylpiperazin-1-yl)phenyl]-N~4~-{[(2R)-oxolan-2-yl]methyl}pyrimidine-2,4-diamine, Activated CDC42 kinase 1, CHLORIDE ION | | Authors: | Thakur, M.K, Miller, W.T, Mahajan, N, Seeliger, M.A. | | Deposit date: | 2020-11-10 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Inhibiting ACK1-mediated phosphorylation of C-terminal Src kinase counteracts prostate cancer immune checkpoint blockade resistance.
Nat Commun, 13, 2022
|
|
2M5E
 
 | | Structure of the C-domain of Calcium-saturated Calmodulin bound to the IQ motif of NaV1.2 | | Descriptor: | CALCIUM ION, Calmodulin, Sodium channel protein type 2 subunit alpha | | Authors: | Fowler, C.A, Feldkamp, M.D, Yu, L, Shea, M.A. | | Deposit date: | 2013-02-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Calcium triggers reversal of calmodulin on nested anti-parallel sites in the IQ motif of the neuronal voltage-dependent sodium channel NaV1.2.
Biophys. Chem., 224, 2017
|
|
5WT7
 
 | | FAS1-IV domain of Human Periostin | | Descriptor: | Periostin | | Authors: | Yun, H, Lee, C.W. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | 1H,13C, and 15N resonance assignments of FAS1-IV domain of human periostin, a component of extracellular matrix proteins.
Biomol NMR Assign, 12, 2018
|
|
4L4Y
 
 | |
4L4Z
 
 | | Crystal structures of the LsrR proteins complexed with phospho-AI-2 and its two different analogs reveal distinct mechanisms for ligand recognition | | Descriptor: | (2S)-2,3,3-trihydroxy-4-oxopentyl dihydrogen phosphate, Transcriptional regulator LsrR | | Authors: | Ryu, K.S, Ha, J.H, Eo, Y. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the LsrR Proteins Complexed with Phospho-AI-2 and Two Signal-Interrupting Analogues Reveal Distinct Mechanisms for Ligand Recognition.
J.Am.Chem.Soc., 135, 2013
|
|
