1DMQ
 
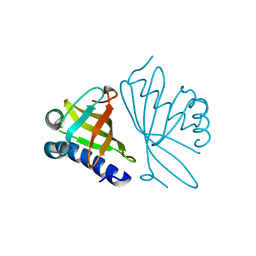 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMN
 
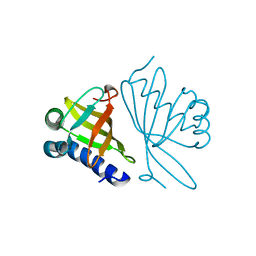 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F/Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMM
 
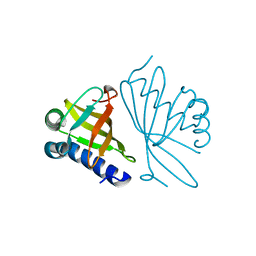 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
7EWI
 
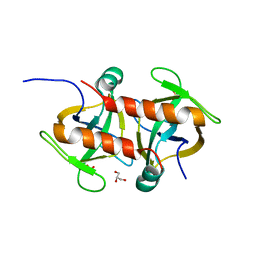 | | Toxin protein from Staphylococcus aureus | | Descriptor: | Endoribonuclease MazF, GLYCEROL, PHOSPHATE ION | | Authors: | Kim, D.H, Kang, S.M, Lee, S.J, Lee, B.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of PemI in the Staphylococcus aureus PemIK toxin-antitoxin complex: PemI controls PemK by acting as a PemK loop mimic.
Nucleic Acids Res., 50, 2022
|
|
7EWJ
 
 | | Toxin-antitoxin complex from Staphylococcus aureus | | Descriptor: | Endoribonuclease MazF, GLYCEROL, PemI inhibitor, ... | | Authors: | Kim, D.H, Kang, S.M, Lee, S.J, Lee, B.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of PemI in the Staphylococcus aureus PemIK toxin-antitoxin complex: PemI controls PemK by acting as a PemK loop mimic.
Nucleic Acids Res., 50, 2022
|
|
8H0H
 
 | |
8H2D
 
 | |
1C7H
 
 | | CRYSTAL STRUCTURE OF A MUTANT R75A IN KETOSTEROID ISOMERASE FROM PSEDOMONAS PUTIDA BIOTYPE B | | Descriptor: | DELTA-5-3-KETOSTEROID ISOMERASE | | Authors: | Nam, G.H, Kim, D.H, Jang, D.S, Choi, G, Ha, N.C, Oh, B.H, Choi, K.Y. | | Deposit date: | 2000-02-19 | | Release date: | 2000-04-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Roles of active site aromatic residues in catalysis by ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 38, 1999
|
|
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
5YRZ
 
 | | Toxin-Antitoxin complex from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, HicA, HicB, ... | | Authors: | Kang, S.M, Kim, D.H. | | Deposit date: | 2017-11-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Functional insights into the Streptococcus pneumoniae HicBA toxin-antitoxin system based on a structural study.
Nucleic Acids Res., 46, 2018
|
|
4XSJ
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4XTB
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4WQ0
 
 | |
5BZ6
 
 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4FXB
 
 | | Crystal structure of CYP105N1 from Streptomyces coelicolor: a cytochrome P450 oxidase in the coelibactin siderophore biosynthetic pathway | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Hong, M.K, Lim, Y.R, Kim, J.K, Kim, D.H, Kang, L.W. | | Deposit date: | 2012-07-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of cytochrome P450 CYP105N1 from Streptomyces coelicolor, an oxidase in the coelibactin siderophore biosynthetic pathway
Arch.Biochem.Biophys., 528, 2012
|
|
7WWP
 
 | | Crystal structure of human Npl4 | | Descriptor: | Nuclear protein localization protein 4 homolog, ZINC ION | | Authors: | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
7WWQ
 
 | | Crystal structure of human Ufd1-Npl4 complex | | Descriptor: | Nuclear protein localization protein 4 homolog, Ubiquitin recognition factor in ER-associated degradation protein 1 | | Authors: | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
4J7A
 
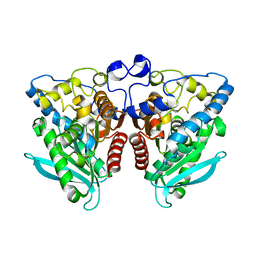 | | Crystal Structure of Est25 - a Bacterial Homolog of Hormone-Sensitive Lipase from a Metagenomic Library | | Descriptor: | Esterase | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Park, K.S, Joo, S.B, Kim, K.K, Kim, D.H. | | Deposit date: | 2013-02-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Structural and functional analyses of a bacterial homologue of hormone-sensitive lipase from a metagenomic library
Acta Crystallogr.,Sect.D, 69, 2014
|
|
6K1G
 
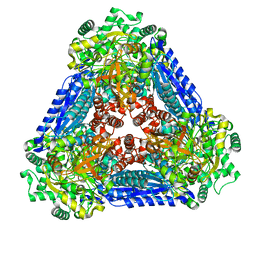 | | Crystal structure of the L-fucose isomerase soaked with Mn2+ from Raoultella sp. | | Descriptor: | L-fucose isomerase, MANGANESE (II) ION | | Authors: | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
6K1F
 
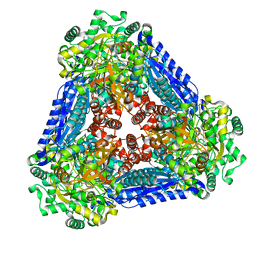 | | Crystal structure of the L-fucose isomerase from Raoultella sp. | | Descriptor: | L-fucose isomerase, MANGANESE (II) ION | | Authors: | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
1CQS
 
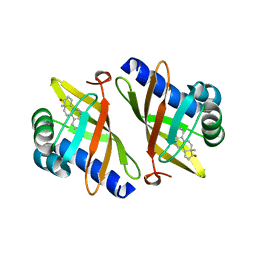 | | CRYSTAL STRUCTURE OF D103E MUTANT WITH EQUILENINEOF KSI IN PSEUDOMONAS PUTIDA | | Descriptor: | EQUILENIN, PROTEIN : KETOSTEROID ISOMERASE | | Authors: | Choi, G, Ha, N.C, Kim, S.W, Kim, D.H, Park, S, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-08-11 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Asp-99 donates a hydrogen bond not to Tyr-14 but to the steroid directly
in the catalytic mechanism of Delta 5-3-ketosteroid isomerase from
Pseudomonas putida biotype B
Biochemistry, 39, 2000
|
|
6JG0
 
 | | Crystal structure of the N-terminal domain single mutant (S92E) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Endolysin,Calcium uniporter protein, SULFATE ION | | Authors: | Lee, Y, Park, J, Min, C.K, Kang, J.Y, Kim, T.G, Yamamoto, T, Kim, D.H, Eom, S.H. | | Deposit date: | 2019-02-13 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of calcium channel domain
To Be Published
|
|
1IY7
 
 | | Crystal Structure of CPA and sulfamide-based inhibitor complex | | Descriptor: | Carboxypeptidase A, PHENYLALANINE-N-SULFONAMIDE, ZINC ION | | Authors: | Kim, S.J, Woo, J.R, Park, J.D, Kim, D.H, Ryu, S.E. | | Deposit date: | 2002-07-24 | | Release date: | 2003-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfamide-Based Inhibitors for Carboxypeptidase A. Novel Type Transition State Analogue Inhibitors for Zinc Proteases
J.Med.Chem., 45, 2002
|
|
1IE6
 
 | | SOLUTION STRUCTURE OF IMPERATOXIN A | | Descriptor: | IMPERATOXIN A | | Authors: | Lee, C.W, Takeuchi, K, Takahashi, H, Sato, K, Shimada, I, Kim, D.H, Kim, J.I. | | Deposit date: | 2001-04-07 | | Release date: | 2003-06-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A.
Biochem.J., 377, 2004
|
|
1HDU
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with aminocarbonylphenylalanine at 1.75 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(AMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
