8HY5
 
 | | Structure of D-amino acid oxidase mutant R38H | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, D-amino-acid oxidase, ... | | Authors: | Khan, S, Upadhyay, S, Dave, U, Kumar, A, Gomes, J. | | Deposit date: | 2023-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into ALS patient derived mutations in D-amino acid oxidase.
Int.J.Biol.Macromol., 256, 2023
|
|
3IRK
 
 | | Solution Structure of Heparin dp30 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
3IRI
 
 | | Solution Structure of Heparin dp18 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
3IRJ
 
 | | Solution Structure of Heparin dp24 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
3IRL
 
 | | Solution Structure of Heparin dp36 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
6FXB
 
 | | Bovine beta-lactoglobulin variant A at pH 4.0 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Major allergen beta-lactoglobulin, NITRATE ION | | Authors: | Khan, S, Ipsen, R, Almdal, K, Svensson, B, Harris, P. | | Deposit date: | 2018-03-08 | | Release date: | 2018-05-23 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revealing the Dimeric Crystal and Solution Structure of beta-Lactoglobulin at pH 4 and Its pH and Salt Dependent Monomer-Dimer Equilibrium.
Biomacromolecules, 19, 2018
|
|
4H02
 
 | |
4JFA
 
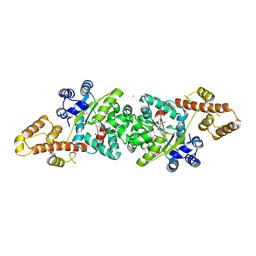 | | Crystal Structure of Plasmodium falciparum Tryptophanyl-tRNA synthetase | | Descriptor: | BETA-MERCAPTOETHANOL, POTASSIUM ION, TRYPTOPHAN, ... | | Authors: | Khan, S, Garg, A, Manickam, Y, Sharma, A. | | Deposit date: | 2013-02-28 | | Release date: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An appended domain results in an unusual architecture for malaria parasite tryptophanyl-tRNA synthetase
Plos One, 8, 2013
|
|
3SZE
 
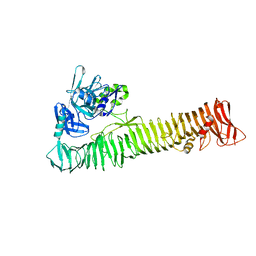 | | Crystal structure of the passenger domain of the E. coli autotransporter EspP | | Descriptor: | Serine protease espP | | Authors: | Khan, S, Mian, H.S, Sandercock, L.E, Battaile, K.P, Lam, R, Chirgadze, N.Y, Pai, E.F. | | Deposit date: | 2011-07-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Passenger Domain of the Escherichia coli Autotransporter EspP.
J.Mol.Biol., 413, 2011
|
|
4LNS
 
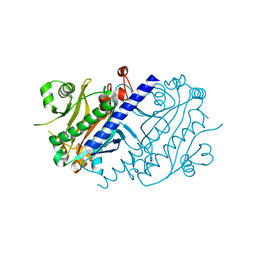 | |
4PG3
 
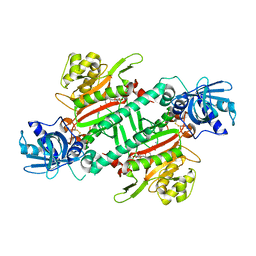 | | Crystal structure of KRS complexed with inhibitor | | Descriptor: | LYSINE, Lysine--tRNA ligase, cladosporin | | Authors: | Sharma, A, Yogavel, M, Khan, S, Sharma, A, Belrhali, H. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin.
J. Struct. Funct. Genomics, 15, 2014
|
|
6P3Q
 
 | | Calpain-5 (CAPN5) Protease Core (PC) | | Descriptor: | Calpain-5 | | Authors: | Velez, G, Sun, Y.J, Khan, S, Yang, J, Koster, H.J, Lokesh, G, Mahajan, V. | | Deposit date: | 2019-05-24 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Unique Activation Mechanisms of a Non-classical Calpain and Its Disease-Causing Variants.
Cell Rep, 30, 2020
|
|
7S16
 
 | | Crystal structure of alpha-COP-WD40 domain R57A mutant | | Descriptor: | Coatomer subunit alpha, SODIUM ION | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7S22
 
 | | Crystal structure of alpha-COP-WD40 domain | | Descriptor: | Coatomer subunit alpha | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-02 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7S23
 
 | | Crystal structure of alpha-COP-WD40 domain, Y139A mutant | | Descriptor: | Coatomer subunit alpha | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
3LMU
 
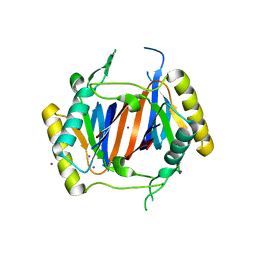 | | Crystal structure of DTD from Plasmodium falciparum | | Descriptor: | D-tyrosyl-tRNA(Tyr) deacylase, IODIDE ION | | Authors: | Manickam, Y, Bhatt, T.K, Khan, S, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3LMT
 
 | | Crystal structure of DTD from Plasmodium falciparum | | Descriptor: | D-tyrosyl-tRNA(Tyr) deacylase, IODIDE ION | | Authors: | Manickam, Y, Bhatt, T.K, Khan, S, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3LMV
 
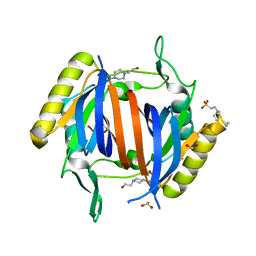 | | D-Tyr-tRNA(Tyr) Deacylase from plasmodium falciparum in complex with hepes | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-tyrosyl-tRNA(Tyr) deacylase, SULFITE ION | | Authors: | Manickam, Y, Khan, S, Bhatt, T.K, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4OO9
 
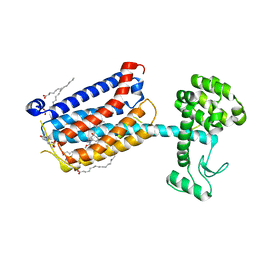 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator mavoglurant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mavoglurant, Metabotropic glutamate receptor 5, ... | | Authors: | Dore, A.S, Okrasa, K, Patel, J.C, Serrano-Vega, M, Bennett, K, Cooke, R.M, Errey, J.C, Jazayeri, A, Khan, S, Tehan, B, Weir, M, Wiggin, G.R, Marshall, F.H. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain.
Nature, 511, 2014
|
|
6JL3
 
 | |
3VGJ
 
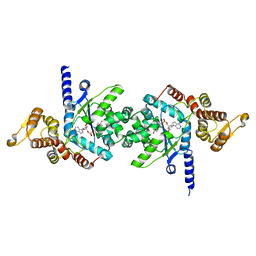 | | Crystal of Plasmodium falciparum tyrosyl-tRNA synthetase (PfTyrRS)in complex with adenylate analog | | Descriptor: | ADENOSINE MONOPHOSPHATE, TYROSINE, Tyrosyl-tRNA synthetase, ... | | Authors: | Banday, M.M, Yogavel, M, Bhatt, T.K, Khan, S, Sharma, A, Sharma, A. | | Deposit date: | 2011-08-14 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Malaria parasite tyrosyl-tRNA synthetase secretion triggers pro-inflammatory responses.
Nat Commun, 2, 2011
|
|
8V2Y
 
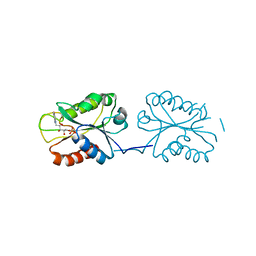 | | Room temperature X-ray Crystal Structure of FMN-bound long-chain flavodoxin from Rhodopseudomonas palustris | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Ansari, A, Khan, S.A, Miller, A.F. | | Deposit date: | 2023-11-24 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure, dynamics, and redox reactivity of an all-purpose flavodoxin.
J.Biol.Chem., 300, 2024
|
|
8SNZ
 
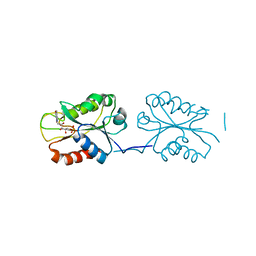 | | X-ray Crystal Structure of FMN-bound long-chain flavodoxin from Rhodopseudomonas palustris | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Ansari, A, Khan, S.A, Miller, A.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure, dynamics, and redox reactivity of an all-purpose flavodoxin.
J.Biol.Chem., 300, 2024
|
|
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
