1KCQ
 
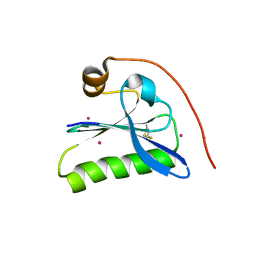 | | Human Gelsolin Domain 2 with a Cd2+ bound | | Descriptor: | CADMIUM ION, GELSOLIN | | Authors: | Kazmirski, S.L, Isaacson, R.L, An, C, Buckle, A, Johnson, C.M, Daggett, V, Fersht, A.R. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Loss of a metal-binding site in gelsolin leads to familial amyloidosis-Finnish type.
Nat.Struct.Biol., 9, 2002
|
|
1XXI
 
 | | ADP Bound E. coli Clamp Loader Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit gamma, DNA polymerase III, ... | | Authors: | Kazmirski, S.L, Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XXH
 
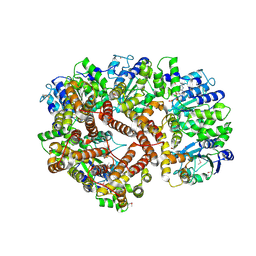 | | ATPgS Bound E. Coli Clamp Loader Complex | | Descriptor: | DNA polymerase III subunit gamma, DNA polymerase III, delta prime subunit, ... | | Authors: | Kazmirski, S.L, Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5BNM
 
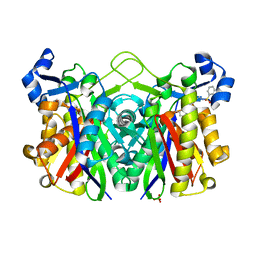 | | E. coli FabH with Small Molecule Inhibitor 1 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, N-{[3'-(hydroxymethyl)biphenyl-4-yl]methyl}benzenesulfonamide, SULFATE ION, ... | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5BNR
 
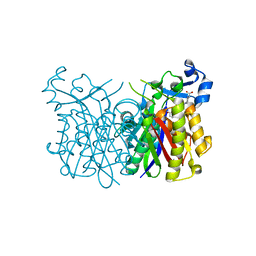 | | E. coli Fabh with small molecule inhibitor 2 | | Descriptor: | 1-{5-[2-fluoro-5-(hydroxymethyl)phenyl]pyridin-2-yl}piperidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5BQS
 
 | | S. Pneumoniae Fabh with small molecule inhibitor 4 | | Descriptor: | 1-{5-[2-chloro-5-(hydroxymethyl)phenyl]pyridin-2-yl}piperidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3, SODIUM ION | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-29 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5BNS
 
 | | E. coli Fabh with small molecule inhibitor 2 | | Descriptor: | 1-{5-[2-fluoro-5-(hydroxymethyl)phenyl]pyridin-2-yl}-N-(quinolin-6-ylmethyl)piperidine-4-carboxamide, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
3GLH
 
 | | Crystal Structure of the E. coli clamp loader bound to Psi Peptide | | Descriptor: | DNA polymerase III subunit delta, DNA polymerase III subunit delta', DNA polymerase III subunit tau | | Authors: | Kazmirski, S.L, Simonetta, K.R, Kuriyan, J. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.891 Å) | | Cite: | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
3K3I
 
 | | p38alpha bound to novel DGF-out compound PF-00215955 | | Descriptor: | (3S)-3-[4-(4-bromophenyl)-1H-imidazol-2-yl]-1,2,3,4-tetrahydroisoquinoline, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Kazmirski, S.L, DiNitto, J.P. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The design, synthesis and potential utility of fluorescence probes that target DFG-out conformation of p38alpha for high throughput screening binding assay.
Chem.Biol.Drug Des., 74, 2009
|
|
3K3J
 
 | | P38alpha bound to novel DFG-out compound PF-00416121 | | Descriptor: | 2-(4-fluorophenyl)-3-oxo-6-pyridin-4-yl-N-[2-(trifluoromethyl)benzyl]-2,3-dihydropyridazine-4-carboxamide, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Kazmirski, S.L, DiNitto, J.P. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | The Design, Synthesis and Potential Utility of Fluorescence Probes that Target DFG-out Conformation of p38alpha for High Throughput Screening Binding Assay.
Chem.Biol.Drug Des., 74, 2009
|
|
4W6E
 
 | | Human Tankyrase 1 with small molecule inhibitor | | Descriptor: | 2-(4-{6-[(3S)-3,4-dimethylpiperazin-1-yl]-4-methylpyridin-3-yl}phenyl)-8-(hydroxymethyl)quinazolin-4(3H)-one, Tankyrase-1, ZINC ION | | Authors: | Kazmirski, S.L, Johannes, J, Boriack-Sjodin, P.A, Howard, T. | | Deposit date: | 2014-08-20 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology.
Acs Med.Chem.Lett., 6, 2015
|
|
5ECE
 
 | | Tankyrase 1 with Phthalazinone 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[3-[(4-oxidanylidene-3~{H}-phthalazin-1-yl)methyl]phenyl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, ACETATE ION, ... | | Authors: | Kazmirski, S.L, Johannes, J, Read, J.A, Howard, T, Larsen, N.A, Ferguson, A.D. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5EBT
 
 | | Tankyrase 1 with Phthalazinone 2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-[bis(fluoranyl)-[3-[[(6~{S})-6-methyl-3-(trifluoromethyl)-6,8-dihydro-5~{H}-[1,2,4]triazolo[4,3-a]pyrazin-7-yl]carbonyl]phenyl]methyl]-2~{H}-phthalazin-1-one, Tankyrase-1, ... | | Authors: | Kazmirski, S.L, Johannes, J. | | Deposit date: | 2015-10-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3DV3
 
 | | MEK1 with PF-04622664 Bound | | Descriptor: | 3,4-difluoro-2-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)-5-[(2-hydroxyethoxy)methyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Kazmirski, S.L, Kothe, M, Ding, Y.-H. | | Deposit date: | 2008-07-18 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Beyond the MEK-pocket: can current MEK kinase inhibitors be utilized to synthesize novel type III NCKIs? Does the MEK-pocket exist in kinases other than MEK?
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4W5S
 
 | | Tankyrase in complex with compound | | Descriptor: | 8-(hydroxymethyl)-2-[4-(1-methyl-1H-pyrazol-4-yl)phenyl]quinazolin-4(3H)-one, GLYCEROL, Tankyrase-1, ... | | Authors: | Johannes, J, Kazmirski, S.L, Boriack-Sjodin, P.A, Howard, T. | | Deposit date: | 2014-08-18 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology.
Acs Med.Chem.Lett., 6, 2015
|
|
6QB3
 
 | | Apo Mcl1 in a complex with a scFv | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, scFv55 | | Authors: | Kazmirski, S, Hargreaves, D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1EM8
 
 | | Crystal structure of chi and psi subunit heterodimer from DNA POL III | | Descriptor: | DNA POLYMERASE III CHI SUBUNIT, DNA POLYMERASE III PSI SUBUNIT | | Authors: | Gulbis, J.M, Finkelstein, J, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2000-03-16 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex.
Eur.J.Biochem., 271, 2004
|
|
6QF9
 
 | | Structure of an anti-Mcl1 scFv | | Descriptor: | scFv | | Authors: | Luptak, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-11-06 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QFC
 
 | | Structure of an anti-Mcl1 scFv | | Descriptor: | DIMETHYL SULFOXIDE, Induced myeloid leukemia cell differentiation protein Mcl-1, scFv55 | | Authors: | Luptak, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-11-06 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6FS1
 
 | | MCL1 in complex with an indole acid ligand | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-[(1,5-dimethylpyrazol-3-yl)methylsulfanylmethyl]-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Kasmirski, S, Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
6FS2
 
 | | MCL1 in complex with indole acid ligand | | Descriptor: | 7-(2-methylphenyl)-3-[3-(5,6,7,8-tetrahydronaphthalen-1-yloxy)propyl]-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
6FS0
 
 | |
4KNR
 
 | | Hin GlmU bound to WG188 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, N-{4-[(2-benzyl-7-hydroxy-6-methoxyquinazolin-4-yl)amino]phenyl}benzamide, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-10 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
4KPZ
 
 | | Hin GlmU bound to a small molecule fragment | | Descriptor: | 1-(3-nitrophenyl)dihydropyrimidine-2,4(1H,3H)-dione, Bifunctional protein GlmU, MAGNESIUM ION, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-14 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
4KNX
 
 | | Hin GlmU Bound to WG176 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-10 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
