1GIF
 
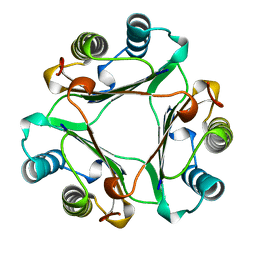 | | HUMAN GLYCOSYLATION-INHIBITING FACTOR | | Descriptor: | GLYCOSYLATION-INHIBITING FACTOR | | Authors: | Kato, Y, Kuroki, R. | | Deposit date: | 1996-02-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human glycosylation-inhibiting factor is a trimeric barrel with three 6-stranded beta-sheets.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1ZR7
 
 | |
1LF8
 
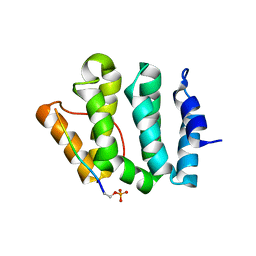 | | Complex of GGA3-VHS Domain and CI-MPR C-terminal Phosphopeptide | | Descriptor: | ADP-ribosylation factor binding protein GGA3, Cation-independent mannose-6-phosphate receptor | | Authors: | Kato, Y, Misra, S, Puertollano, R, Hurley, J.H, Bonifacino, J.S. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phosphoregulation of sorting signal-VHS domain interactions by a direct electrostatic mechanism.
Nat.Struct.Biol., 9, 2002
|
|
5ZJ9
 
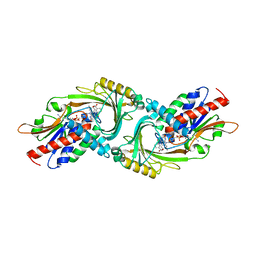 | | human D-amino acid oxidase complexed with 5-chlorothiophene-3-carboxylic acid | | Descriptor: | 5-chloro thiophene-3-carboxylic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kato, Y, Hin, N, Maita, N, Thomas, A.G, Kurosawa, S, Rojas, C, Yorita, K, Slusher, B.S, Fukui, K, Tsukamoto, T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids
Eur J Med Chem, 159, 2018
|
|
5ZJA
 
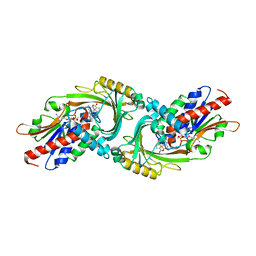 | | human D-amino acid oxidase complexed with 5-chlorothiophene-2-carboxylic acid | | Descriptor: | 5-chloro thiophene-2-carboxylic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kato, Y, Hin, N, Maita, N, Thomas, A.G, Kurosawa, S, Rojas, C, Yorita, K, Slusher, B.S, Fukui, K, Tsukamoto, T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids
Eur J Med Chem, 159, 2018
|
|
8I59
 
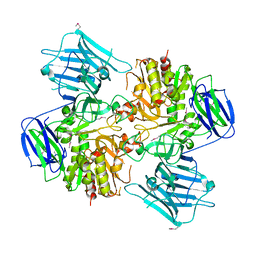 | | N-acetyl-(R)-beta-phenylalanine acylase, selenomethionyl derivative | | Descriptor: | N-acetyl-(R)-beta-phenylalanine acylase | | Authors: | Kato, Y, Natsume, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Expression, purification and crystallization of N-acetyl-(R)-beta-phenylalanine acylases derived from Burkholderia sp. AJ110349 and Variovorax sp. AJ110348 and structure determination of the Burkholderia enzyme.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8I5A
 
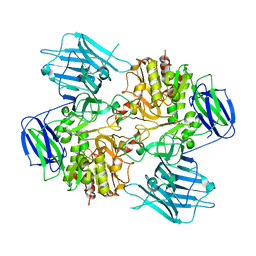 | | N-acetyl-(R)-beta-phenylalanine acylase, 2.75 angstrom resolution | | Descriptor: | N-acetyl-(R)-beta-phenylalanine acylase | | Authors: | Kato, Y, Natsume, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Expression, purification and crystallization of N-acetyl-(R)-beta-phenylalanine acylases derived from Burkholderia sp. AJ110349 and Variovorax sp. AJ110348 and structure determination of the Burkholderia enzyme.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8HUY
 
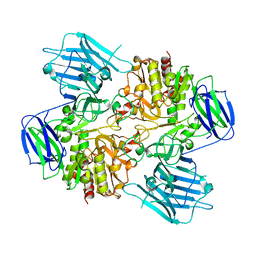 | | N-acetyl-(R)-beta-phenylalanine acylase | | Descriptor: | Chains: A,B | | Authors: | Kato, Y, Natsume, R. | | Deposit date: | 2022-12-24 | | Release date: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expression, purification and crystallization of N-acetyl-(R)-beta-phenylalanine acylases derived from Burkholderia sp. AJ110349 and Variovorax sp. AJ110348 and structure determination of the Burkholderia enzyme.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
2DYF
 
 | |
6L3S
 
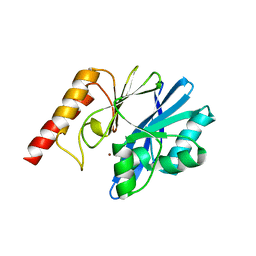 | |
1IV9
 
 | |
1IV7
 
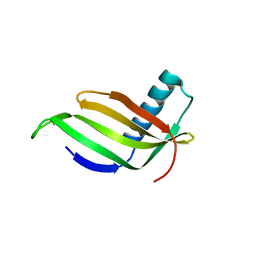 | |
1EHA
 
 | | CRYSTAL STRUCTURE OF GLYCOSYLTREHALOSE TREHALOHYDROLASE FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Authors: | Feese, M.D, Kato, Y, Tamada, T, Kato, M, Komeda, T, Kobayashi, K, Kuroki, R. | | Deposit date: | 2000-02-19 | | Release date: | 2001-02-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus.
J.Mol.Biol., 301, 2000
|
|
1EH9
 
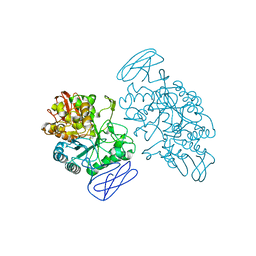 | | CRYSTAL STRUCTURE OF SULFOLOBUS SOLFATARICUS GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Descriptor: | GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Authors: | Feese, M.D, Kato, Y, Tamada, T, Kato, M, Komeda, T, Kobayashi, K, Kuroki, R. | | Deposit date: | 2000-02-19 | | Release date: | 2001-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus.
J.Mol.Biol., 301, 2000
|
|
1V7M
 
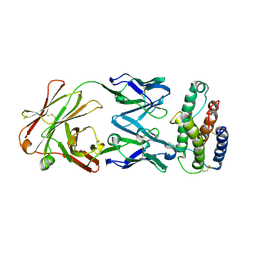 | | Human Thrombopoietin Functional Domain Complexed To Neutralizing Antibody TN1 Fab | | Descriptor: | Monoclonal TN1 Fab Heavy Chain, Monoclonal TN1 Fab Light Chain, Thrombopoietin | | Authors: | Feese, M.D, Tamada, T, Kato, Y, Maeda, Y, Hirose, M, Matsukura, Y, Shigematsu, H, Kato, T, Miyazaki, H, Kuroki, R. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the receptor-binding domain of human thrombopoietin determined by complexation with a neutralizing antibody fragment
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1V7N
 
 | | Human Thrombopoietin Functional Domain Complexed To Neutralizing Antibody TN1 Fab | | Descriptor: | Monoclonal TN1 Fab Heavy Chain, Monoclonal TN1 Fab Light Chain, Thrombopoietin | | Authors: | Feese, M.D, Tamada, T, Kato, Y, Maeda, Y, Hirose, M, Matsukura, Y, Shigematsu, H, Kato, T, Miyazaki, H, Kuroki, R. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the receptor-binding domain of human thrombopoietin determined by complexation with a neutralizing antibody fragment
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1KKO
 
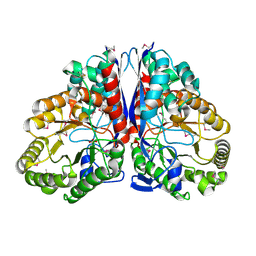 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE | | Descriptor: | 3-METHYLASPARTATE AMMONIA-LYASE, SULFATE ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, Y, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
2JV4
 
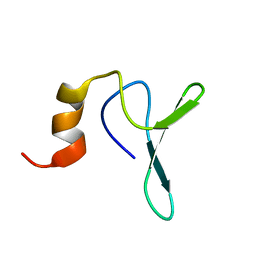 | | Structure Characterisation of PINA WW Domain and Comparison with other Group IV WW Domains, PIN1 and ESS1 | | Descriptor: | Peptidyl-prolyl cis/trans isomerase | | Authors: | Ng, C.A, Kato, Y, Tanokura, M, Brownlee, R.T.C. | | Deposit date: | 2007-09-11 | | Release date: | 2007-10-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural characterisation of PinA WW domain and a comparison with other Group IV WW domains, Pin1 and Ess1
Biochim.Biophys.Acta, 1784, 2008
|
|
6A3L
 
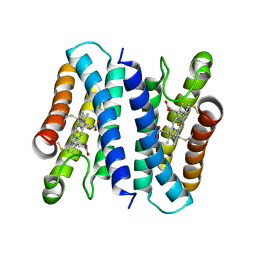 | | Crystal structure of cytochrome c' from Shewanella violacea DSS12 | | Descriptor: | Cytochrome c, HEME C | | Authors: | Suka, A, Oki, H, Kato, Y, Kawahara, K, Ohkubo, T, Maruno, T, Kobayashi, Y, Fujii, S, Wakai, S, Sambongi, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-12 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Stability of cytochromes c' from psychrophilic and piezophilic Shewanella species: implications for complex multiple adaptation to low temperature and high hydrostatic pressure.
Extremophiles, 23, 2019
|
|
6A3K
 
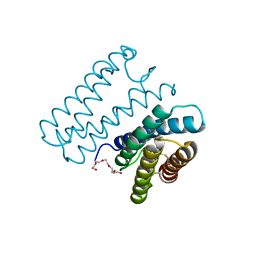 | | Crystal structure of cytochrome c' from Shewanella benthica DB6705 | | Descriptor: | Cytochrome c, HEME C, PENTAETHYLENE GLYCOL | | Authors: | Suka, A, Oki, H, Kato, Y, Kawahara, K, Ohkubo, T, Maruno, T, Kobayashi, Y, Fujii, S, Wakai, S, Sambongi, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-12 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Stability of cytochromes c' from psychrophilic and piezophilic Shewanella species: implications for complex multiple adaptation to low temperature and high hydrostatic pressure.
Extremophiles, 23, 2019
|
|
5GIS
 
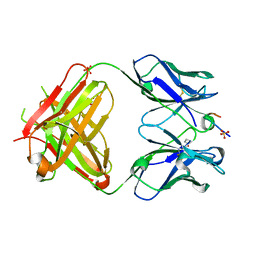 | | Crystal structure of a Fab fragment with its ligand peptide | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, SULFATE ION, ... | | Authors: | Kitago, Y, Kaneko, K.K, Ogasawara, S, Kato, Y, Takagi, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for multi-specific peptide recognition by the anti-IDH1/2 monoclonal antibody, MsMab-1.
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GIR
 
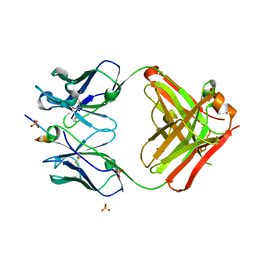 | | Crystal structure of a Fab fragment with its ligand peptide | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of Fab fragment, LYS-PRO-ILE-ILE-ILE-GLY-SER-HIS-ALA-TYR-GLY-ASP, ... | | Authors: | Kitago, Y, Kaneko, K.K, Ogasawara, S, Kato, Y, Takagi, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for multi-specific peptide recognition by the anti-IDH1/2 monoclonal antibody, MsMab-1.
Biochem. Biophys. Res. Commun., 478, 2016
|
|
1IUQ
 
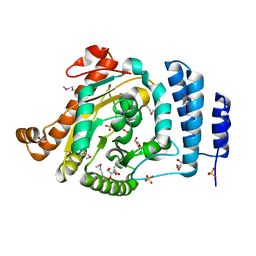 | | The 1.55 A Crystal Structure of Glycerol-3-Phosphate Acyltransferase | | Descriptor: | GLYCEROL, Glycerol-3-Phosphate Acyltransferase, SULFATE ION | | Authors: | Tamada, T, Feese, M.D, Kato, Y, Kuroki, R. | | Deposit date: | 2002-03-06 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate recognition and selectivity of plant glycerol-3-phosphate acyltransferases (GPATs) from Cucurbita moscata and Spinacea oleracea.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3WWK
 
 | | Crystal structure of CLEC-2 in complex with rhodocytin | | Descriptor: | C-type lectin domain family 1 member B, Snaclec rhodocytin subunit alpha, Snaclec rhodocytin subunit beta | | Authors: | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | Deposit date: | 2014-06-20 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
3WSR
 
 | | Crystal structure of CLEC-2 in complex with O-glycosylated podoplanin | | Descriptor: | C-type lectin domain family 1 member B, Peptide from Podoplanin, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | Deposit date: | 2014-03-20 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
