6IU9
 
 | | Crystal structure of cytoplasmic metal binding domain with iron ions | | Descriptor: | FE (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU5
 
 | | Crystal structure of cytoplasmic metal binding domain with zinc ions | | Descriptor: | CHLORIDE ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU4
 
 | | Crystal structure of iron transporter VIT1 with cobalt ion | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU8
 
 | | Crystal structure of cytoplasmic metal binding domain with cobalt ions | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU3
 
 | | Crystal structure of iron transporter VIT1 with zinc ions | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU6
 
 | | Crystal structure of cytoplasmic metal binding domain with nickel ions | | Descriptor: | NICKEL (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
7VR7
 
 | | Inward-facing structure of human EAAT2 in the WAY213613-bound state | | Descriptor: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-22 | | Release date: | 2022-08-10 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7VR8
 
 | | Inward-facing structure of human EAAT2 in the substrate-free state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-22 | | Release date: | 2022-08-10 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
6K9Q
 
 | | Structure of the native supercoiled hook as a universal joint | | Descriptor: | Flagellar hook protein FlgE | | Authors: | Kato, T, Miyata, T, Makino, F, Horvath, P, Namba, K. | | Deposit date: | 2019-06-17 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the native supercoiled flagellar hook as a universal joint.
Nat Commun, 10, 2019
|
|
7WLS
 
 | | Crystal structure of the multidrug efflux transporter BpeB from Burkholderia pseudomallei | | Descriptor: | Efflux pump membrane transporter, TETRAETHYLENE GLYCOL, UNDECYL-MALTOSIDE | | Authors: | Kato, T, Hung, L.-W, Yamashita, E, Okada, U, Terwilliger, T.C, Murakami, S. | | Deposit date: | 2022-01-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structures of multidrug efflux transporters from Burkholderia pseudomallei suggest details of transport mechanism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WLV
 
 | | Crystal Structure of the Multidrug effulx transporter BpeF from Burkholderia pseudomallei. | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Kato, T, Hung, L.-W, Yamashita, E, Okada, U, Terwilliger, T.C, Murakami, S. | | Deposit date: | 2022-01-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of multidrug efflux transporters from Burkholderia pseudomallei suggest details of transport mechanism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6O97
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with propylamycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.75A resolution | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxyc yclohexyl 2-amino-2,4-dideoxy-4-propyl-alpha-D-glucopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Matsushita, T, Sati, G.C, Kondasinghe, N, Pirrone, M.G, Kato, T, Waduge, P, Kumar, H.S, Sanchon, A.C, Dobosz-Bartoszek, M, Shcherbakov, D, Juhas, M, Hobbie, S.N, Schrepfer, T, Chow, C.S, Polikanov, Y.S, Schacht, J, Vasella, A, Bottger, E.C, Crich, D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Multigram Synthesis, and in Vitro and in Vivo Evaluation of Propylamycin: A Semisynthetic 4,5-Deoxystreptamine Class Aminoglycoside for the Treatment of Drug-Resistant Enterobacteriaceae and Other Gram-Negative Pathogens.
J. Am. Chem. Soc., 141, 2019
|
|
8GQY
 
 | | CryoEM structure of pentameric MotA from Aquifex aeolicus | | Descriptor: | Motility protein A | | Authors: | Nishikino, T, Takekawa, N, Kishikawa, J, Hirose, M, Onoe, S, Kato, T, Imada, K. | | Deposit date: | 2022-08-31 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of MotA, a flagellar stator protein, from hyperthermophile.
Biochem.Biophys.Res.Commun., 631, 2022
|
|
3J0R
 
 | | Model of a type III secretion system needle based on a 7 Angstrom resolution cryoEM map | | Descriptor: | Protein mxiH | | Authors: | Fujii, T, Cheung, M, Blanco, A, Kato, T, Blocker, A.J, Namba, K. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of a type III secretion needle at 7-A resolution provides insights into its assembly and signaling mechanisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4D3E
 
 | | Tetramer of IpaD, modified from 2J0O, fitted into negative stain electron microscopy reconstruction of the wild type tip complex from the type III secretion system of Shigella flexneri | | Descriptor: | INVASIN IPAD | | Authors: | Cheung, M, Shen, D.-K, Makino, F, Kato, T, Roehrich, D, Martinez-Argudo, I, Walker, M.L, Murillo, I, Liu, X, Pain, M, Brown, J, Frazer, G, Mantell, J, Mina, P, Todd, T, Sessions, R.B, Namba, K, Blocker, A.J. | | Deposit date: | 2014-10-21 | | Release date: | 2014-12-10 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Three-Dimensional Electron Microscopy Reconstruction and Cysteine-Mediated Crosslinking Provide a Model of the T3Ss Needle Tip Complex.
Mol.Microbiol., 95, 2015
|
|
1RCH
 
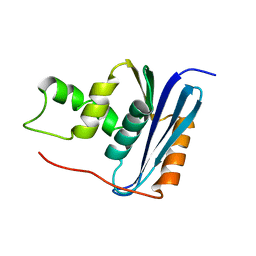 | | SOLUTION NMR STRUCTURE OF RIBONUCLEASE HI FROM ESCHERICHIA COLI, 8 STRUCTURES | | Descriptor: | RIBONUCLEASE HI | | Authors: | Yamazaki, T, Fujiwara, M, Kato, T, Yamasaki, K, Nagayama, K. | | Deposit date: | 1995-06-23 | | Release date: | 1997-02-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribonuclease Hi from Escherichia Coli
Biol.Pharm.Bull., 23, 2000
|
|
8BVS
 
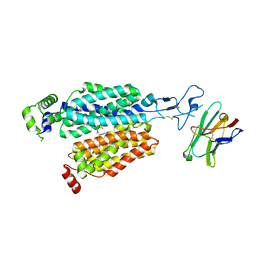 | | Cryo-EM structure of rat SLC22A6 bound to tenofovir | | Descriptor: | CHLORIDE ION, Solute carrier family 22 member 6, Synthetic nanobody (Sybody), ... | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-05 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BVT
 
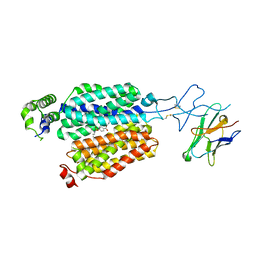 | | Cryo-EM structure of rat SLC22A6 bound to probenecid | | Descriptor: | 4-(dipropylsulfamoyl)benzoic acid, Solute carrier family 22 member 6, Synthetic nanobody (Sybody) | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-06 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BW7
 
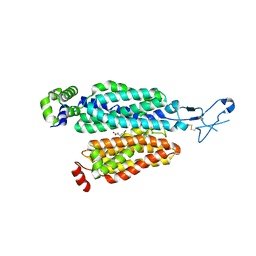 | | Cryo-EM structure of rat SLC22A6 bound to alpha-ketoglutaric acid | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, Solute carrier family 22 member 6, ... | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-06 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BVR
 
 | | Cryo-EM structure of rat SLC22A6 in the apo state | | Descriptor: | PHOSPHATE ION, Solute carrier family 22 member 6, Synthetic nanobody (Sybody) | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-05 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1V7M
 
 | | Human Thrombopoietin Functional Domain Complexed To Neutralizing Antibody TN1 Fab | | Descriptor: | Monoclonal TN1 Fab Heavy Chain, Monoclonal TN1 Fab Light Chain, Thrombopoietin | | Authors: | Feese, M.D, Tamada, T, Kato, Y, Maeda, Y, Hirose, M, Matsukura, Y, Shigematsu, H, Kato, T, Miyazaki, H, Kuroki, R. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the receptor-binding domain of human thrombopoietin determined by complexation with a neutralizing antibody fragment
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1V7N
 
 | | Human Thrombopoietin Functional Domain Complexed To Neutralizing Antibody TN1 Fab | | Descriptor: | Monoclonal TN1 Fab Heavy Chain, Monoclonal TN1 Fab Light Chain, Thrombopoietin | | Authors: | Feese, M.D, Tamada, T, Kato, Y, Maeda, Y, Hirose, M, Matsukura, Y, Shigematsu, H, Kato, T, Miyazaki, H, Kuroki, R. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the receptor-binding domain of human thrombopoietin determined by complexation with a neutralizing antibody fragment
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8JJE
 
 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
8HLB
 
 | | Cryo-EM structure of biparatopic antibody Bp109-92 in complex with TNFR2 | | Descriptor: | TR109 heavy chain, TR109 light chain, TR92 heavy chain, ... | | Authors: | Akiba, H, Fujita, J, Ise, T, Nishiyama, K, Miyata, T, Kato, T, Namba, K, Ohno, H, Kamada, H, Nagata, S, Tsumoto, K. | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Development of a 1:1-binding biparatopic anti-TNFR2 antagonist by reducing signaling activity through epitope selection.
Commun Biol, 6, 2023
|
|
7C86
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: Dark state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RETINAL, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
