3AMO
 
 | | Time-resolved X-ray Crystal Structure Analysis of Enzymatic Reaction of Copper Amine Oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Kataoka, M, Oya, H, Tominaga, A, Otsu, M, Okajima, T, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2010-08-20 | | Release date: | 2011-11-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detection of the reaction intermediates catalyzed by a copper amine oxidase.
J.SYNCHROTRON RADIAT., 18, 2011
|
|
3WQ0
 
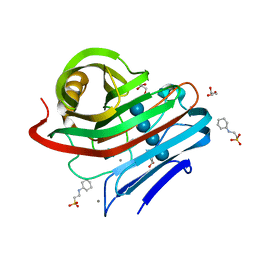 | |
3WQ1
 
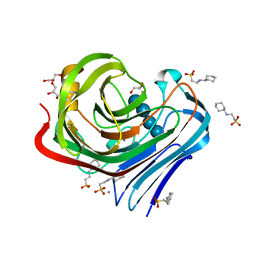 | |
3WR0
 
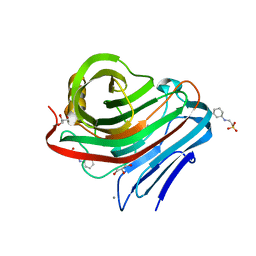 | |
3WQ7
 
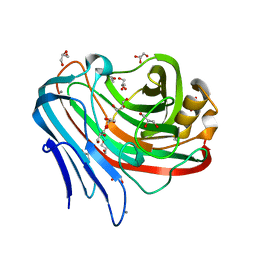 | |
3WXP
 
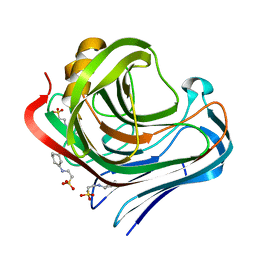 | |
3WT3
 
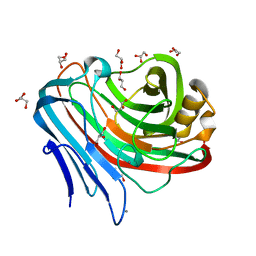 | |
3WY6
 
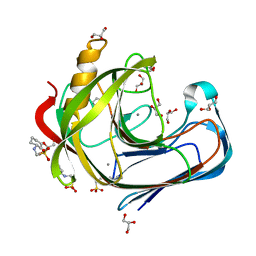 | |
3VGI
 
 | | The crystal structure of hyperthermophilic family 12 endocellulase from Pyrococcus furiosus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Endoglucanase A, ... | | Authors: | Kataoka, M, Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-08-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic resolution of the crystal structure of the hyperthermophilic family 12 endocellulase and stabilizing role of the DxDxDG calcium-binding motif in Pyrococcus furiosus.
Febs Lett., 586, 2012
|
|
3WP3
 
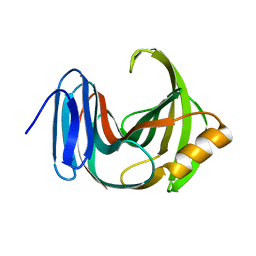 | |
4YPJ
 
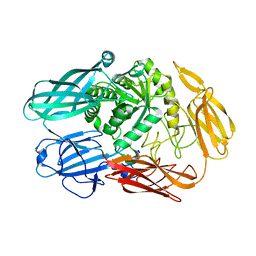 | | X-ray Structure of The Mutant of Glycoside Hydrolase | | Descriptor: | Beta galactosidase | | Authors: | Ishikawa, K, Kataoka, M, Yanamoto, T, Nakabayashi, M, Watanabe, M. | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of beta-galactosidase from Bacillus circulans ATCC 31382 (BgaD) and the construction of the thermophilic mutants.
Febs J., 282, 2015
|
|
4TMC
 
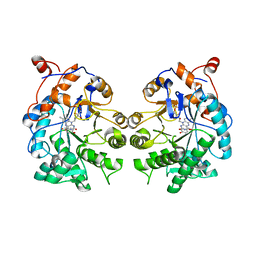 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 COMPLEXED with P-HYDROXYBENZALDEHYDE | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme, P-HYDROXYBENZALDEHYDE | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
4TMB
 
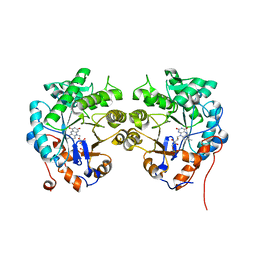 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
2D01
 
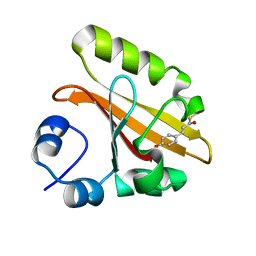 | | Wild Type Photoactive Yellow Protein, P65 Form | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Shimizu, N, Kamikubo, H, Yamazaki, Y, Imamoto, Y, Kataoka, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The Crystal Structure of the R52Q Mutant Demonstrates a Role for R52 in Chromophore pK(a) Regulation in Photoactive Yellow Protein
Biochemistry, 45, 2006
|
|
2D02
 
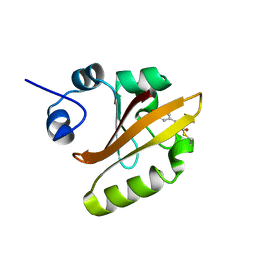 | | R52Q Mutant of Photoactive Yellow Protein, P65 Form | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Shimizu, N, Kamikubo, H, Yamazaki, Y, Imamoto, Y, Kataoka, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Crystal Structure of the R52Q Mutant Demonstrates a Role for R52 in Chromophore pK(a) Regulation in Photoactive Yellow Protein
Biochemistry, 45, 2006
|
|
6AGZ
 
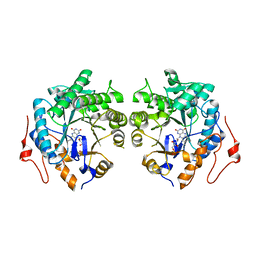 | | Crystal structure of Old Yellow Enzyme from Pichia sp. AKU4542 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old Yellow Enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2018-08-15 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of different substrate preferences of two old yellow enzymes from yeasts in the asymmetric reduction of enone compounds.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
5B36
 
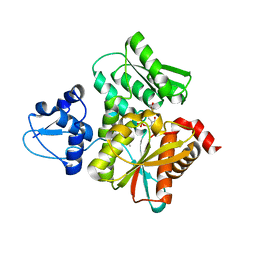 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with Cysteine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
5B3A
 
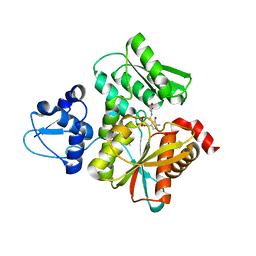 | | Crystal Structure of O-Phoshoserine Sulfhydrylase from Aeropyrum pernix in Complexed with the alpha-Aminoacrylate Intermediate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Protein CysO | | Authors: | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2016-02-12 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
2ZAM
 
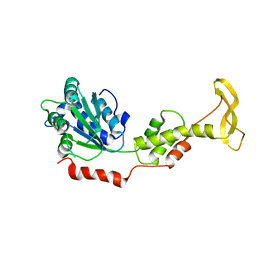 | | Crystal structure of mouse SKD1/VPS4B apo-form | | Descriptor: | Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZAO
 
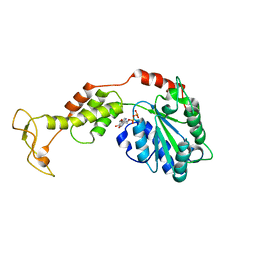 | | Crystal structure of mouse SKD1/VPS4B ADP-form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZAN
 
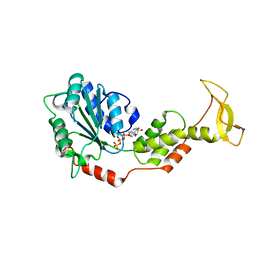 | | Crystal structure of mouse SKD1/VPS4B ATP-form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
3AK4
 
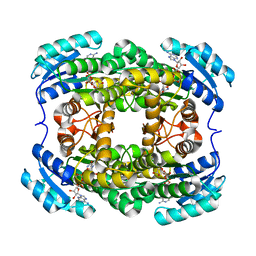 | | Crystal structure of NADH-dependent quinuclidinone reductase from agrobacterium tumefaciens | | Descriptor: | NADH-dependent quinuclidinone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miyakawa, T, Kataoka, M, Takeshita, D, Nomoto, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NADH-dependent quinuclidinone reductase from Agrobacterium tumefaciens
To be Published
|
|
1V4R
 
 | | Solution structure of Streptmycal repressor TraR | | Descriptor: | Transcriptional Repressor | | Authors: | Tanaka, T, Komatsu, C, Kobayashi, K, Sugai, M, Kataoka, M, Kohno, T. | | Deposit date: | 2003-11-17 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Streptmycal repressor TraR
TO BE PUBLISHED
|
|
2IEZ
 
 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2IEY
 
 | | Crystal Structure of mouse Rab27b bound to GDP in hexagonal space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
