4HX9
 
 | | Designed Phosphodeoxyribosyltransferase | | Descriptor: | Nucleoside deoxyribosyltransferase, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Kaminski, P.A, Labesse, G. | | Deposit date: | 2012-11-09 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Phosphodeoxyribosyltransferases, designed enzymes for deoxyribonucleotides synthesis.
J.Biol.Chem., 288, 2013
|
|
2KLH
 
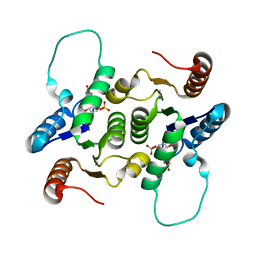 | | NMR Structure of RCL in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, c-Myc-responsive protein Rcl | | Authors: | Padilla, A, Yang, Y, Labesse, G, Zhang, C, Kaminski, P.A. | | Deposit date: | 2009-07-02 | | Release date: | 2009-10-20 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the mammalian deoxynucleotide N-hydrolase Rcl and its stabilizing interactions with two inhibitors
J.Mol.Biol., 394, 2009
|
|
6RM2
 
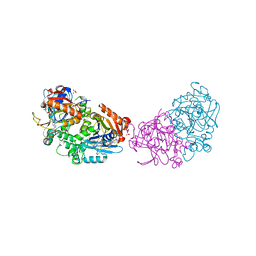 | | Deoxyguanylosuccinate synthase (DgsS) structure with ATP, IMP, Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, INOSINIC ACID, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-05-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP, IMP, Magnesium at 2.5 Angstrom resolution
To Be Published
|
|
6TNH
 
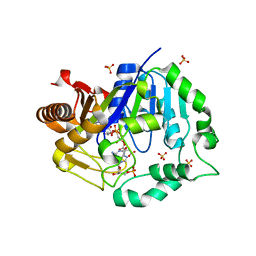 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with AMPPcP, dGMP, Asp, Magnesium at 2.21 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ASPARTIC ACID, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
4P5E
 
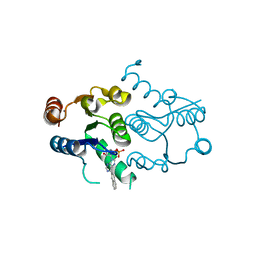 | | CRYSTAL STRUCTURE OF HUMAN DNPH1 (RCL) WITH 6-NAPHTHYL-PURINE-RIBOSIDE-MONOPHOSPHATE | | Descriptor: | 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 6-(naphthalen-2-yl)-9-(5-O-phosphono-beta-D-ribofuranosyl)-9H-purine, CALCIUM ION | | Authors: | Padilla, A, Labesse, G, Kaminski, P.A. | | Deposit date: | 2014-03-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: Synthesis, structural studies and cytotoxic activities.
Eur.J.Med.Chem., 85C, 2014
|
|
4P5D
 
 | | CRYSTAL STRUCTURE OF RAT DNPH1 (RCL) WITH 6-NAPHTHYL-PURINE-RIBOSIDE-MONOPHOSPHATE | | Descriptor: | 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 6-(naphthalen-2-yl)-9-(5-O-phosphono-beta-D-ribofuranosyl)-9H-purine, SULFATE ION | | Authors: | Padilla, A, Labesse, G, Kaminski, P.A. | | Deposit date: | 2014-03-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: Synthesis, structural studies and cytotoxic activities.
Eur.J.Med.Chem., 85C, 2014
|
|
6FKO
 
 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP, dGMP, hadacidin at 2.1 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-24 | | Release date: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP, dGMP, HAdacidin at 2.1 Angstrom resolution
To Be Published
|
|
6FLF
 
 | |
6FM1
 
 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATPanddGMP at 2.3 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-29 | | Release date: | 2019-06-12 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
6FM0
 
 | | Deoxyguanylosuccinate synthase (DgsS) and ATP structure at 1.7 Angstrom resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-29 | | Release date: | 2019-06-12 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
6FM3
 
 | | Deoxyguanylosuccinate synthase (DgsS) structure with ADP at 1.9 Angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylosuccinate synthetase, CHLORIDE ION | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-30 | | Release date: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP0, dGMP, Magnesium at 2.3 Angstrom resolution
To Be Published
|
|
4KXM
 
 | |
4KXL
 
 | |
4KXN
 
 | |
6ZZA
 
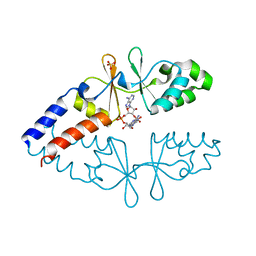 | | Crystal structure of CbpB in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CBS domain-containing protein, SULFATE ION | | Authors: | Mechaly, A.E, Covaleda-Cortes, G, Kaminski, P.A. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of CbpB in complex with c-di-AMP
To Be Published
|
|
6ZZ9
 
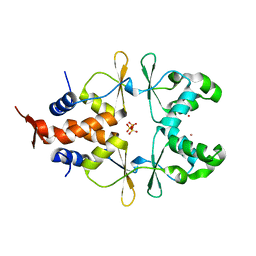 | |
4FYH
 
 | | Crystal structure of rcl with phospho-triciribine | | Descriptor: | 5-methyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-1,5-dihydro-1,4,5,6,8-pentaazaacenaphthylen-3-amine, Deoxyribonucleoside 5'-monophosphate N-glycosidase, SULFATE ION | | Authors: | Labesse, G, Padilla, A, Kaminski, P.A. | | Deposit date: | 2012-07-04 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of the oncoprotein Rcl bound to three nucleotide analogues.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FYK
 
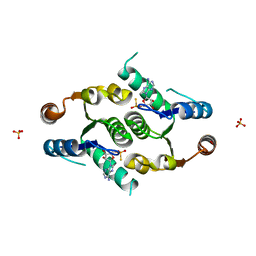 | | Crystal structure of rcl with 5'-phiosphorothioate-adenosine | | Descriptor: | ADENOSINE -5'-THIO-MONOPHOSPHATE, Deoxyribonucleoside 5'-monophosphate N-glycosidase, SULFATE ION | | Authors: | Labesse, G, Padilla, A, Kaminski, P.A. | | Deposit date: | 2012-07-04 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the oncoprotein Rcl bound to three nucleotide analogues.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FYI
 
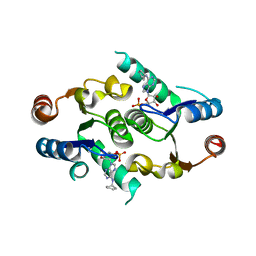 | | Crystal structure of rcl with 6-cyclopentyl-AMP | | Descriptor: | Deoxyribonucleoside 5'-monophosphate N-glycosidase, N-cyclopentyladenosine 5'-(dihydrogen phosphate) | | Authors: | Labesse, G, Padilla, A, Kaminski, P.A. | | Deposit date: | 2012-07-04 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of the oncoprotein Rcl bound to three nucleotide analogues.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JEN
 
 | | Structure of Clostridium botulinum CMP N-glycosidase, BcmB | | Descriptor: | CMP N-GLYCOSIDASE, PHOSPHATE ION | | Authors: | Sikowitz, M.D, Cooper, L.E, Begley, T.P, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reversal of the substrate specificity of CMP N-glycosidase to dCMP.
Biochemistry, 52, 2013
|
|
4JEL
 
 | | Structure of MilB Streptomyces rimofaciens CMP N-glycosidase | | Descriptor: | CMP/hydroxymethyl CMP hydrolase, SULFATE ION | | Authors: | Sikowitz, M.D, Cooper, L.E, Begley, T.P, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Reversal of the substrate specificity of CMP N-glycosidase to dCMP.
Biochemistry, 52, 2013
|
|
4JEM
 
 | | Crystal structure of MilB complexed with cytidine 5'-monophosphate | | Descriptor: | CMP/hydroxymethyl CMP hydrolase, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Sikowitz, M.D, Cooper, L.E, Begley, T.P, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Reversal of the substrate specificity of CMP N-glycosidase to dCMP.
Biochemistry, 52, 2013
|
|
1S2L
 
 | |
1S2G
 
 | | Purine 2'deoxyribosyltransferase + 2'-deoxyadenosine | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, purine trans deoxyribosylase | | Authors: | Anand, R, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1S2I
 
 | | Purine 2'deoxyribosyltransferase + bromopurine | | Descriptor: | 6-BROMO-7H-PURINE, purine trans deoxyribosylase | | Authors: | Anand, R, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
