4WL1
 
 | |
5J96
 
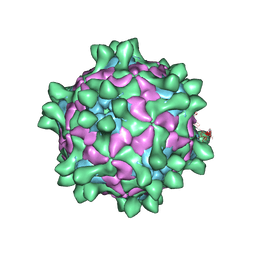 | | Crystal structure of Slow Bee Paralysis Virus at 3.4A resolution | | Descriptor: | Genome polyprotein, VP1, VP2 | | Authors: | Kalynych, S, Levdansky, Y, Palkova, L, Plevka, P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Virion Structure of Iflavirus Slow Bee Paralysis Virus at 2.6-Angstrom Resolution.
J.Virol., 90, 2016
|
|
5J98
 
 | | Crystal structure of Slow Bee Paralysis Virus at 2.6A resolution | | Descriptor: | VP1, VP2, VP3 | | Authors: | Kalynych, S, Levdansky, Y, Palkova, L, Plevka, P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Virion Structure of Iflavirus Slow Bee Paralysis Virus at 2.6-Angstrom Resolution.
J.Virol., 90, 2016
|
|
5LK8
 
 | |
5LK7
 
 | |
4Z92
 
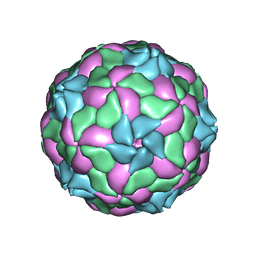 | | crystal structure of parechovirus-1 virion | | Descriptor: | Capsid subunit VP3, RNA (5'-R(*AP*UP*UP*UP*UP*U)-3'), capsid subunit VP0, ... | | Authors: | Kalynych, S, Palkova, L, Plevka, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of Human Parechovirus 1 Reveals an Association of the RNA Genome with the Capsid.
J.Virol., 90, 2015
|
|
4E2C
 
 | |
4E2L
 
 | |
4E2H
 
 | |
4E29
 
 | |
5M48
 
 | |
5CDD
 
 | |
5CDC
 
 | |
5M9D
 
 | |
7OPX
 
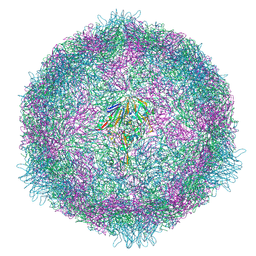 | | CryoEM structure of human enterovirus 70 native virion | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Fuzik, T, Plevka, P, Moravcova, J. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds.
J.Virol., 96, 2022
|
|
7OZK
 
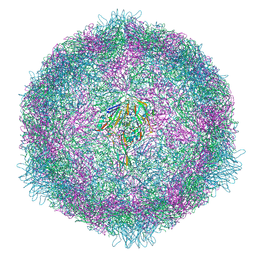 | | CryoEM structure of human enterovirus 70 in complex with Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Fuzik, T, Plevka, P, Moravcova, J. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds.
J.Virol., 96, 2022
|
|
7OZL
 
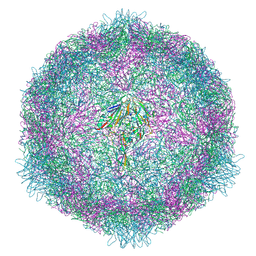 | | CryoEM structure of human enterovirus 70 in complex with WIN51711 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Fuzik, T, Plevka, P, Moravcova, J. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds.
J.Virol., 96, 2022
|
|
7OZJ
 
 | |
7OZI
 
 | |
