4Y9H
 
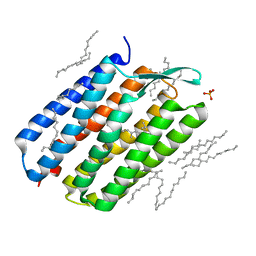 | | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, DECANE, DODECANE, ... | | Authors: | Saiki, H, Sugiyama, S, Kakinouchi, K, Kawatake, S, Hanashima, S, Matsumori, N, Murata, M. | | Deposit date: | 2015-02-17 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles
To Be Published
|
|
7V5U
 
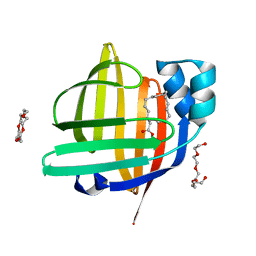 | | The 0.92 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with 2-cyclohexadecylacetic acid (CYC16AA) | | Descriptor: | 2-cyclohexadecylethanoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | The 0.92 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with 2-cyclohexadecylacetic acid (CYC16AA)
To Be Published
|
|
7VB1
 
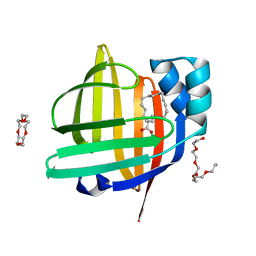 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with trans-vaccenic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-08-30 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with trans-vaccenic acid
To Be Published
|
|
3WXQ
 
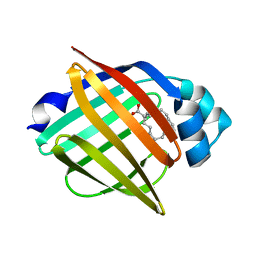 | | Serial femtosecond X-ray structure of human fatty acid-binding protein type-3 (FABP3) in complex with stearic acid (C18:0) determined using X-ray free-electron laser at SACLA | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Mizohata, E, Suzuki, M, Kakinouchi, K, Sugiyama, S, Murata, M, Sugahara, M, Nango, E, Tanaka, T, Tanaka, R, Tono, K, Song, C, Hatsui, T, Joti, Y, Yabashi, M, Iwata, S. | | Deposit date: | 2014-08-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
7WCI
 
 | | The 0.85 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with two molecules of pelargonic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The 0.85 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with two molecules of pelargonic acid
To Be Published
|
|
7WF0
 
 | | The 0.83 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with nervonic acid | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-24 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | The 0.83 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with nervonic acid
To Be Published
|
|
7WJ1
 
 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with arachidonic acid | | Descriptor: | ARACHIDONIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with arachidonic acid
To Be Published
|
|
7WKG
 
 | | The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with erucic acid | | Descriptor: | (Z)-docos-13-enoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with erucic acid
To Be Published
|
|
7WKB
 
 | | The 0.81 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with docosahexaenoic acid | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-08 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | The 0.81 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with docosahexaenoic acid
To Be Published
|
|
7WOM
 
 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with eicosapentaenoic acid | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Fatty acid-binding protein, heart | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with eicosapentaenoic acid
To Be Published
|
|
5B27
 
 | | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | Authors: | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid.
To Be Published
|
|
5B29
 
 | | The 1.28A structure of human FABP3 F16V mutant complexed with palmitic acid at room temperature | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID | | Authors: | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The 1.28A structure of human FABP3 F16V mutant complexed with palmitic acid at room temperature.
To Be Published
|
|
5B28
 
 | | The 0.90A structure of human FABP3 F16V mutant complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | Authors: | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The 0.90A structure of human FABP3 F16V mutant complexed with palmitic acid.
To Be Published
|
|
7FD7
 
 | | The 1.00 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluoroheptanoic acid | | Descriptor: | Fatty acid-binding protein, heart, PENTAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Hara, T, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The 1.00 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluoroheptanoic acid
To Be Published
|
|
7FDT
 
 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with elaidic acid | | Descriptor: | 9-OCTADECENOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-17 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with octanoic acid
To Be Published
|
|
7FDU
 
 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with heptadecanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with heptadecanoic acid
To Be Published
|
|
7FCX
 
 | | Room temperature structure of the human heart fatty acid-binding protein complexed with hexanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXANOIC ACID | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Room temperature structure of the human heart fatty acid-binding protein complexed with hexanoic acid
To Be Published
|
|
7FCG
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing Indometacin | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing Indometacin
To Be Published
|
|
7FF6
 
 | | The 0.83 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with cis-vaccenic acid | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | The 0.83 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with cis-vaccenic acid
To Be Published
|
|
7FEU
 
 | | The 0.95 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluorononanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Hara, T, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The 0.95 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluorononanoic acid
To Be Published
|
|
7FEZ
 
 | | The 0.76 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with petroselinic acid | | Descriptor: | Fatty acid-binding protein, heart, PENTAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.76 Å) | | Cite: | The 0.76 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with petroselinic acid
To Be Published
|
|
7FFK
 
 | | The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with palmitoleic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-23 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with palmitoleic acid
To Be Published
|
|
7FEK
 
 | | The 1.05 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluorooctanoic acid | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Hara, T, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The 1.05 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluorooctanoic acid
To Be Published
|
|
7FG5
 
 | | Room temperature structure of the human heart fatty acid-binding protein complexed with capric acid | | Descriptor: | DECANOIC ACID, Fatty acid-binding protein, heart | | Authors: | Sugiyama, S, Kakinouchi, K, Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-26 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Room temperature structure of the human heart fatty acid-binding protein complexed with capric acid
To Be Published
|
|
7EUW
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing 4-[2-[1-(4-bromophenyl)-5-phenyl-1H-pyrazol-3-yl]phenoxy] (HA174) | | Descriptor: | 4-[2-[1-(4-bromophenyl)-5-phenyl-pyrazol-3-yl]phenoxy]butanoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-05-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with 4-[2-[1-(4-bromophenyl)-5-phenyl-1H-pyrazol-3-yl]phenoxy] (HA176)
To Be Published
|
|
