1F9C
 
 | | CRYSTAL STRUCTURE OF MLE D178N VARIANT | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE I) | | Authors: | Kajander, T, Lehtio, L, Kahn, P.C, Goldman, A. | | Deposit date: | 2000-07-10 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Buried charged surface in proteins.
Structure Fold.Des., 8, 2000
|
|
8CEG
 
 | |
1JOF
 
 | | Neurospora crassa 3-carboxy-cis,cis-mucoante lactonizing enzyme | | Descriptor: | BETA-MERCAPTOETHANOL, CARBOXY-CIS,CIS-MUCONATE CYCLASE, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Kajander, T, Merckel, M.C, Thompson, A, Deacon, A.M, Mazur, P, Kozarich, J.W, Goldman, A. | | Deposit date: | 2001-07-28 | | Release date: | 2002-04-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Neurospora crassa 3-carboxy-cis,cis-muconate lactonizing enzyme, a beta propeller cycloisomerase.
Structure, 10, 2002
|
|
3ESK
 
 | |
2FO7
 
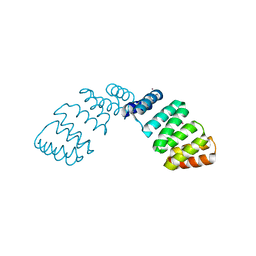 | |
2AVP
 
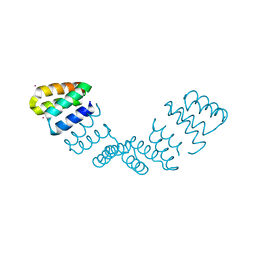 | | Crystal structure of an 8 repeat consensus TPR superhelix | | Descriptor: | CADMIUM ION, synthetic consensus TPR protein | | Authors: | Kajander, T, Cortajarena, A.L, Main, E.R, Mochrie, S, Regan, L. | | Deposit date: | 2005-08-30 | | Release date: | 2005-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure and stability of designed TPR protein superhelices: unusual crystal packing and implications for natural TPR proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2HYZ
 
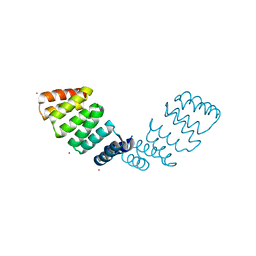 | |
1NU5
 
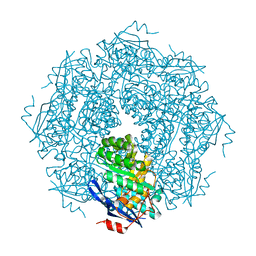 | |
2XOT
 
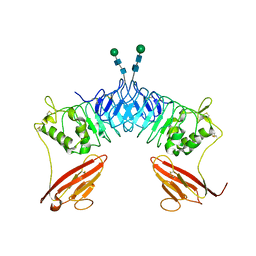 | | Crystal structure of neuronal leucine rich repeat protein AMIGO-1 | | Descriptor: | Amphoterin-induced protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kajander, T, Kuja-Panula, J, Rauvala, H, Goldman, A. | | Deposit date: | 2010-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Role of Glycans and Dimerisation in Folding of Neuronal Leucine-Rich Repeat Protein Amigo-1
J.Mol.Biol., 413, 2011
|
|
2XQW
 
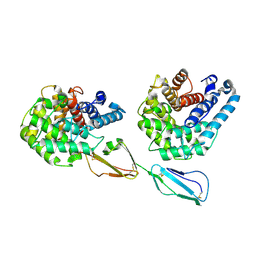 | | Structure of Factor H domains 19-20 in complex with complement C3d | | Descriptor: | COMPLEMENT C3, COMPLEMENT FACTOR H | | Authors: | Kajander, T, Lehtinen, M.J, Hyvarinen, S, Bhattacharjee, A, Leung, E, Isenman, D.E, Meri, S, Jokiranta, T.S, Goldman, A. | | Deposit date: | 2010-09-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Dual Interaction of Factor H with C3D and Glycosaminoglycans in Host-Nonhost Discrimination by Complement.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4AV6
 
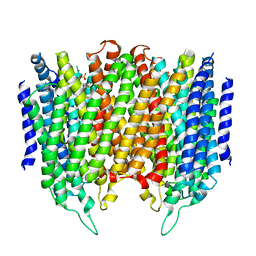 | | Crystal structure of Thermotoga maritima sodium pumping membrane integral pyrophosphatase at 4 A in complex with phosphate and magnesium | | Descriptor: | K(+)-STIMULATED PYROPHOSPHATE-ENERGIZED SODIUM PUMP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kajander, T, Kellosalo, J, Kogan, K, Pokharel, K, Goldman, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Structure and Catalytic Cycle of a Sodium-Pumping Pyrophosphatase.
Science, 337, 2012
|
|
4AV3
 
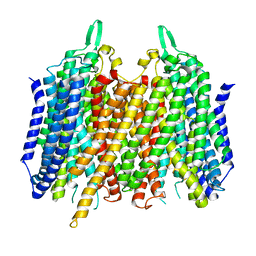 | | Crystal structure of Thermotoga Maritima sodium pumping membrane integral pyrophosphatase with metal ions in active site | | Descriptor: | CALCIUM ION, K(+)-STIMULATED PYROPHOSPHATE-ENERGIZED SODIUM PUMP, MAGNESIUM ION | | Authors: | Kajander, T, Kogan, K, Kellosalo, J, Pokharel, K, Goldman, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure and Catalytic Cycle of a Sodium-Pumping Pyrophosphatase.
Science, 337, 2012
|
|
117E
 
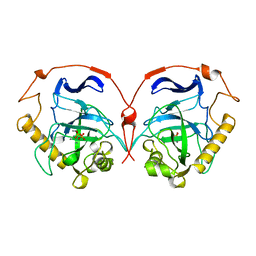 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
8PRK
 
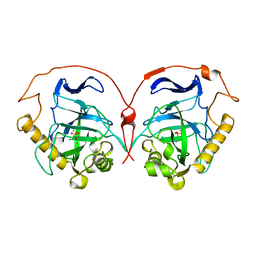 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-16 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
6TL8
 
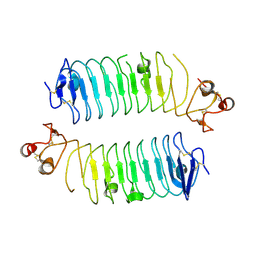 | | Structural basis of SALM3 dimerization and adhesion complex formation with the presynaptic receptor protein tyrosine phosphatases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Myeloid cell surface antigen CD33,Leucine-rich repeat and fibronectin type-III domain-containing protein 4 | | Authors: | Karki, S, Shkumatov, A.V, Bae, S, Ko, J, Kajander, T. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of SALM3 dimerization and synaptic adhesion complex formation with PTP sigma.
Sci Rep, 10, 2020
|
|
6F2O
 
 | |
3MUC
 
 | | MUCONATE CYCLOISOMERASE VARIANT I54V | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE) | | Authors: | Schell, U, Helin, S, Kajander, T, Schlomann, M, Goldman, A. | | Deposit date: | 1998-10-27 | | Release date: | 1999-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates.
Proteins, 34, 1999
|
|
5A5C
 
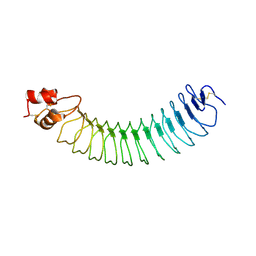 | | Structure of an engineered neuronal LRRTM2 adhesion molecule | | Descriptor: | LRRTM | | Authors: | Paatero, A, Rosti, K, Shkumatov, A.V, Brunello, C, Kysenius, K, Huttunen, H, Kajander, T. | | Deposit date: | 2015-06-17 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal Structure of an Engineered Lrrtm2 Synaptic Adhesion Molecule and a Model for Neurexin Binding.
Biochemistry, 55, 2016
|
|
5LZR
 
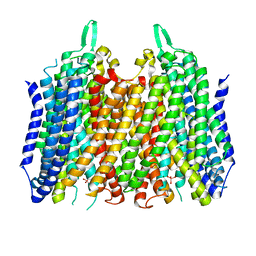 | | Crystal structure of Thermotoga maritima sodium pumping membrane integral pyrophosphatase in complex with tungstate and magnesium | | Descriptor: | K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, TUNGSTATE(VI)ION | | Authors: | Wilkinson, C, Kellosalo, J, Kajander, T, Goldman, A. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Membrane pyrophosphatases from Thermotoga maritima and Vigna radiata suggest a conserved coupling mechanism.
Nat Commun, 7, 2016
|
|
5LZQ
 
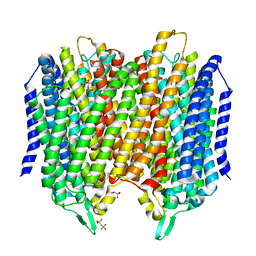 | | Crystal structure of Thermotoga maritima sodium pumping membrane integral pyrophosphatase in complex with imidodiphosphate and magnesium, and with bound sodium ion | | Descriptor: | IMIDODIPHOSPHORIC ACID, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Wilkinson, C, Kellosalo, J, Kajander, T, Goldman, A. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Membrane pyrophosphatases from Thermotoga maritima and Vigna radiata suggest a conserved coupling mechanism.
Nat Commun, 7, 2016
|
|
3J2J
 
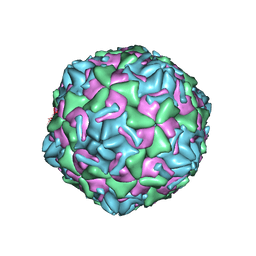 | | Empty coxsackievirus A9 capsid | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Shakeel, S, Seitsonen, J.J.T, Kajander, T, Laurinmaki, P, Hyypia, T, Susi, P, Butcher, S.J. | | Deposit date: | 2012-10-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.54 Å) | | Cite: | Structural and functional analysis of coxsackievirus A9 integrin {alpha}v{beta}6 binding and uncoating.
J.Virol., 87, 2013
|
|
1XNF
 
 | | Crystal structure of E.coli TPR-protein NlpI | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Lipoprotein nlpI | | Authors: | Wilson, C.G, Kajander, T, Regan, L. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of NlpI. A prokaryotic tetratricopeptide repeat protein with a globular fold.
FEBS J., 272, 2005
|
|
5E4V
 
 | | Crystal structure of measles N0-P complex | | Descriptor: | Nucleoprotein,Phosphoprotein | | Authors: | Guryanov, S.G, Liljeroos, L, Kasaragod, P, Kajander, T, Butcher, S.J. | | Deposit date: | 2015-10-07 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of the Measles Virus Nucleoprotein Core in Complex with an N-Terminal Region of Phosphoprotein.
J.Virol., 90, 2015
|
|
4O1S
 
 | | Crystal structure of TvoVMA intein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Aranko, A.S, Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7008 Å) | | Cite: | Structure-based engineering and comparison of novel split inteins for protein ligation.
Mol Biosyst, 10, 2014
|
|
4O1R
 
 | | Crystal structure of NpuDnaB intein | | Descriptor: | CHLORIDE ION, GLYCEROL, Replicative DNA helicase, ... | | Authors: | Aranko, A.S, Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based engineering and comparison of novel split inteins for protein ligation.
Mol Biosyst, 10, 2014
|
|
