3HYJ
 
 | | Crystal structure of the N-terminal LAGLIDADG domain of DUF199/WhiA | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DUF199/WhiA, ... | | Authors: | Kaiser, B.K, Clifton, M.C, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease.
Structure, 17, 2009
|
|
3HYI
 
 | | Crystal structure of full-length DUF199/WhiA from Thermatoga maritima | | Descriptor: | GLYCEROL, Protein DUF199/WhiA, SODIUM ION | | Authors: | Kaiser, B.K, Clifton, M.C, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease
Structure, 17, 2009
|
|
3CII
 
 | | Structure of NKG2A/CD94 bound to HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen peptide, HLA class I histocompatibility antigen, ... | | Authors: | Strong, R.K, Kaiser, B.K, Pizarro, J.C. | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.41 Å) | | Cite: | Structural basis for NKG2A/CD94 recognition of HLA-E.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4YIS
 
 | | Crystal Structure of LAGLIDADG Meganuclease I-CpaMI Bound to Uncleaved DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (28-MER), ... | | Authors: | Hallinan, J.P, Kaiser, B.K, Stoddard, B.L. | | Deposit date: | 2015-03-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity.
Structure, 24, 2016
|
|
4YHX
 
 | |
5E63
 
 | | K262A mutant of I-SmaMI | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-METHOXYETHANOL, DNA (5'-D(P*CP*AP*GP*GP*TP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Shen, B, Stoddard, B. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
5E5P
 
 | | Wild type I-SmaMI in the space group of C121 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, I-SmaMI LAGLIDADG meganuclease | | Authors: | Shen, B.W. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
5E5S
 
 | | I-SmaMI K103A mutant | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Bottom strand DNA, DNA (5'-D(P*CP*AP*GP*GP*TP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Shen, B.W. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
5E5O
 
 | |
5E67
 
 | | K103A/K262A double mutant of I-SmaMI | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-METHOXYETHANOL, DNA bottom strand, ... | | Authors: | Shen, B.W, Stoddard, B. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
5ESP
 
 | |
7T8L
 
 | | BrxR from Acinetobacter BREX type I phage restriction system | | Descriptor: | 1,2-ETHANEDIOL, BrxR, CHLORIDE ION | | Authors: | Doyle, L, Kaiser, B, Stoddard, B. | | Deposit date: | 2021-12-16 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of the WYL BrxR protein and its gene as separable regulatory elements of a BREX phage restriction system.
Nucleic Acids Res., 50, 2022
|
|
7T8K
 
 | | BrxR from Acinetobacter BREX type I phage restriction system bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, BrxR, CHLORIDE ION, ... | | Authors: | Doyle, L, Kaiser, B, Stoddard, B. | | Deposit date: | 2021-12-16 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and characterization of the WYL BrxR protein and its gene as separable regulatory elements of a BREX phage restriction system.
Nucleic Acids Res., 50, 2022
|
|
3U0D
 
 | |
3U9P
 
 | | Crystal Structure of Murine Siderocalin in Complex with an Fab Fragment | | Descriptor: | Monoclonal Fab Fragment Heavy Chain, Monoclonal Fab Fragment Light Chain, Neutrophil gelatinase-associated lipocalin | | Authors: | Correnti, C, Strong, R.K. | | Deposit date: | 2011-10-19 | | Release date: | 2013-05-01 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Siderocalin/Lcn2/NGAL/24p3 does not drive apoptosis through gentisic acid mediated iron withdrawal in hematopoietic cell lines.
Plos One, 7, 2012
|
|
8EIL
 
 | | C-Terminal Domain of BrxL from Acinetobacter BREX type I phage restriction system | | Descriptor: | MALONIC ACID, Protease Lon-related BREX system protein BrxL, SUCCINIC ACID | | Authors: | Doyle, L.A, Stoddard, B.L, Kaiser, B. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure, substrate binding and activity of a unique AAA+ protein: the BrxL phage restriction factor.
Nucleic Acids Res., 51, 2023
|
|
4YIT
 
 | |
4Z20
 
 | |
4Z1Z
 
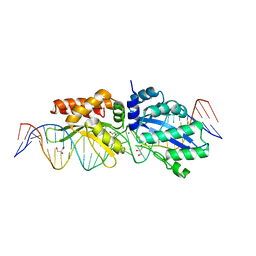 | |
7JTA
 
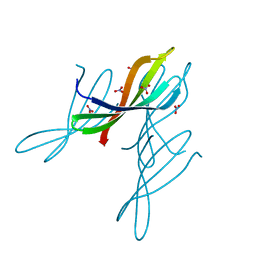 | | Crystal structure of a putative nuclease with anti-Cas9 activity from an uncultured Clostridia bacterium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NITRATE ION, NTF2-like nuclease/anti-CRISPR | | Authors: | Werther, R, Forsberg, K.J, Stoddard, B.L. | | Deposit date: | 2020-08-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The novel anti-CRISPR AcrIIA22 relieves DNA torsion in target plasmids and impairs SpyCas9 activity.
Plos Biol., 19, 2021
|
|
8EMH
 
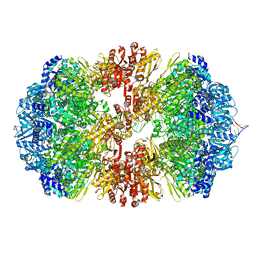 | |
8EMC
 
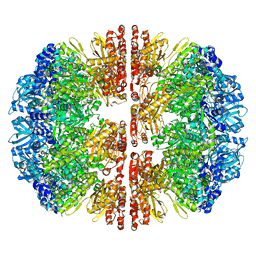 | |
