1BG9
 
 | | BARLEY ALPHA-AMYLASE WITH SUBSTRATE ANALOGUE ACARBOSE | | Descriptor: | 1,4-ALPHA-D-GLUCAN GLUCANOHYDROLASE, 4,6-dideoxy-4-{[(1S,4S,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Kadziola, A, Haser, R. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure of a barley alpha-amylase-inhibitor complex: implications for starch binding and catalysis.
J.Mol.Biol., 278, 1998
|
|
1AMY
 
 | |
1ETP
 
 | |
1U9Z
 
 | | Crystal Structure of Phosphoribosyl Diphosphate Synthase Complexed with AMP and Ribose 5-Phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, RIBOSE-5-PHOSPHATE, Ribose-phosphate pyrophosphokinase | | Authors: | Kadziola, A, Johansson, E, Jepsen, C.H, McGuire, J, Larsen, S, Hove-Jensen, B. | | Deposit date: | 2004-08-11 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii
J.Mol.Biol., 354, 2005
|
|
1U9Y
 
 | | Crystal Structure of Phosphoribosyl Diphosphate Synthase from Methanocaldococcus jannaschii | | Descriptor: | Ribose-phosphate pyrophosphokinase | | Authors: | Kadziola, A, Johansson, E, Jepsen, C.H, McGuire, J, Larsen, S, Hove-Jensen, B. | | Deposit date: | 2004-08-11 | | Release date: | 2005-08-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii
J.Mol.Biol., 354, 2005
|
|
1I5E
 
 | |
3G6W
 
 | |
4TS5
 
 | | Sulfolobus solfataricus adenine phosphoribosyltransferase with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Purine phosphoribosyltransferase (GpT-1) | | Authors: | Kadziola, A. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Adenine Phosphoribosyltransferase from Sulfolobus solfataricus Is an Enzyme with Unusual Kinetic Properties and a Crystal Structure that Suggests It Evolved from a 6-Oxopurine Phosphoribosyltransferase.
Biochemistry, 54, 2015
|
|
4TRB
 
 | | Sulfolobus solfataricus adenine phosphoribosyltransferase | | Descriptor: | PHOSPHATE ION, Purine phosphoribosyltransferase (GpT-1) | | Authors: | Kadziola, A. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Adenine Phosphoribosyltransferase from Sulfolobus solfataricus Is an Enzyme with Unusual Kinetic Properties and a Crystal Structure that Suggests It Evolved from a 6-Oxopurine Phosphoribosyltransferase.
Biochemistry, 54, 2015
|
|
4TRC
 
 | | Sulfolobus solfataricus adenine phosphoribosyltransferase with adenine | | Descriptor: | ADENINE, PHOSPHATE ION, Purine phosphoribosyltransferase (GpT-1) | | Authors: | Kadziola, A. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-30 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Adenine Phosphoribosyltransferase from Sulfolobus solfataricus Is an Enzyme with Unusual Kinetic Properties and a Crystal Structure that Suggests It Evolved from a 6-Oxopurine Phosphoribosyltransferase.
Biochemistry, 54, 2015
|
|
4TS7
 
 | | Sulfolobus solfataricus adenine phosphoribosyltransferase with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Purine phosphoribosyltransferase (GpT-1) | | Authors: | Kadziola, A. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Adenine Phosphoribosyltransferase from Sulfolobus solfataricus Is an Enzyme with Unusual Kinetic Properties and a Crystal Structure that Suggests It Evolved from a 6-Oxopurine Phosphoribosyltransferase.
Biochemistry, 54, 2015
|
|
4TWB
 
 | | Sulfolobus solfataricus ribose-phosphate pyrophosphokinase | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ribose-phosphate pyrophosphokinase, SULFATE ION | | Authors: | Kadziola, A. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of dimeric, recombinant Sulfolobus solfataricus phosphoribosyl diphosphate synthase: a bent dimer defining the adenine specificity of the substrate ATP.
Extremophiles, 19, 2015
|
|
1VST
 
 | | Symmetric Sulfolobus solfataricus uracil phosphoribosyltransferase with bound PRPP and GTP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kadziola, A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and kinetic studies of the allosteric transition in Sulfolobus solfataricus uracil phosphoribosyltransferase: Permanent activation by engineering of the C-terminus
J.Mol.Biol., 393, 2009
|
|
1AVA
 
 | | AMY2/BASI PROTEIN-PROTEIN COMPLEX FROM BARLEY SEED | | Descriptor: | BARLEY ALPHA-AMYLASE 2(CV MENUET), BARLEY ALPHA-AMYLASE/SUBTILISIN INHIBITOR, CALCIUM ION | | Authors: | Vallee, F, Kadziola, A, Bourne, Y, Juy, M, Svensson, B, Haser, R. | | Deposit date: | 1997-09-15 | | Release date: | 1999-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Barley alpha-amylase bound to its endogenous protein inhibitor BASI: crystal structure of the complex at 1.9 A resolution.
Structure, 6, 1998
|
|
6F2P
 
 | | Structure of Paenibacillus xanthan lyase to 2.6 A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lo Leggio, L, Kadziola, A. | | Deposit date: | 2017-11-27 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Dynamics of a Promiscuous Xanthan Lyase from Paenibacillus nanensis and the Design of Variants with Increased Stability and Activity.
Cell Chem Biol, 26, 2019
|
|
2FXV
 
 | | Bacillus subtilis Xanthine Phosphoribosyltransferase in Complex with Guanosine 5'-monophosphate (GMP) | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, Xanthine phosphoribosyltransferase | | Authors: | Arent, S, Kadziola, A, Larsen, S, Neuhard, J, Jensen, K.F. | | Deposit date: | 2006-02-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Extraordinary Specificity of Xanthine Phosphoribosyltransferase from Bacillus subtilis Elucidated
by Reaction Kinetics, Ligand Binding, and Crystallography
Biochemistry, 45, 2006
|
|
1OYS
 
 | |
1OYP
 
 | | Crystal Structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis | | Descriptor: | Ribonuclease PH, SULFATE ION | | Authors: | Harlow, L.S, Kadziola, A, Jensen, K.F, Larsen, S. | | Deposit date: | 2003-04-07 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis and implications for its quaternary structure and tRNA binding.
Protein Sci., 13, 2004
|
|
1OYR
 
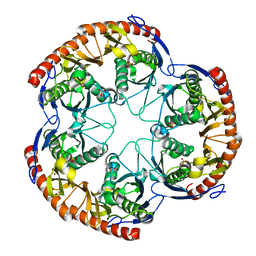 | | Crystal structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis | | Descriptor: | CADMIUM ION, Ribonuclease PH, SULFATE ION | | Authors: | Harlow, L.S, Kadziola, A, Jensen, K.F, Larsen, S. | | Deposit date: | 2003-04-07 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis and implications for its quaternary structure and tRNA binding.
Protein Sci., 13, 2004
|
|
1DKR
 
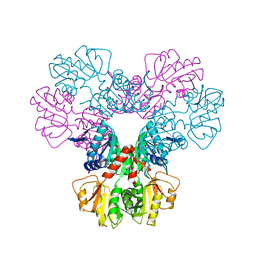 | | CRYSTAL STRUCTURES OF BACILLUS SUBTILIS PHOSPHORIBOSYLPYROPHOSPHATE SYNTHETASE: MOLECULAR BASIS OF ALLOSTERIC INHIBITION AND ACTIVATION. | | Descriptor: | PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Eriksen, T.A, Kadziola, A, Bentsen, A.-K, Harlow, K.W, Larsen, S. | | Deposit date: | 1999-12-08 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.
Nat.Struct.Biol., 7, 2000
|
|
1DKU
 
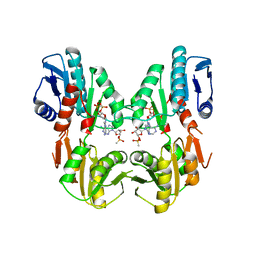 | | CRYSTAL STRUCTURES OF BACILLUS SUBTILIS PHOSPHORIBOSYLPYROPHOSPHATE SYNTHETASE: MOLECULAR BASIS OF ALLOSTERIC INHIBITION AND ACTIVATION. | | Descriptor: | METHYL PHOSPHONIC ACID ADENOSINE ESTER, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PROTEIN (PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE) | | Authors: | Eriksen, T.A, Kadziola, A, Bentsen, A.-K, Harlow, K.W, Larsen, S. | | Deposit date: | 1999-12-08 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.
Nat.Struct.Biol., 7, 2000
|
|
1EK4
 
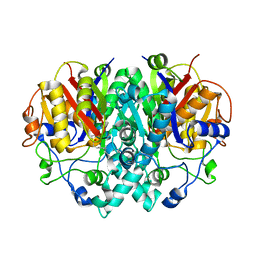 | | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH DODECANOIC ACID TO 1.85 RESOLUTION | | Descriptor: | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I, LAURIC ACID | | Authors: | Olsen, J.G, Kadziola, A, Siggaard-Andersen, M, von Wettstein-Knowles, P, Larsen, S. | | Deposit date: | 2000-03-06 | | Release date: | 2001-04-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of beta-ketoacyl-acyl carrier protein synthase I complexed with fatty acids elucidate its catalytic machinery.
Structure, 9, 2001
|
|
1IBS
 
 | | PHOSPHORIBOSYLDIPHOSPHATE SYNTHETASE IN COMPLEX WITH CADMIUM IONS | | Descriptor: | CADMIUM ION, METHYL PHOSPHONIC ACID ADENOSINE ESTER, RIBOSE-PHOSPHATE PYROPHOSPHOKINASE, ... | | Authors: | Eriksen, T.A, Kadziola, A, Larsen, S. | | Deposit date: | 2001-03-29 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding of cations in Bacillus subtilis phosphoribosyldiphosphate synthetase and their role in catalysis.
Protein Sci., 11, 2002
|
|
1JQP
 
 | | dipeptidyl peptidase I (cathepsin C), a tetrameric cysteine protease of the papain family | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Olsen, J.G, Kadziola, A, Lauritzen, C, Pedersen, J, Larsen, S, Dahl, S.W. | | Deposit date: | 2001-08-08 | | Release date: | 2002-10-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tetrameric dipeptidyl peptidase I directs substrate specificity by use of the residual pro-part domain
FEBS LETT., 506, 2001
|
|
1CPX
 
 | |
