1ATN
 
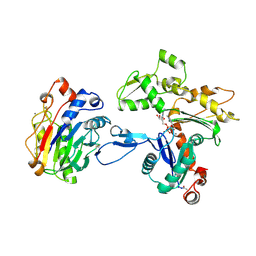 | | Atomic structure of the actin:DNASE I complex | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kabsch, W, Mannherz, H.G, Suck, D, Pai, E, Holmes, K.C. | | Deposit date: | 1991-03-08 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the actin:DNase I complex.
Nature, 347, 1990
|
|
1CLU
 
 | | H-RAS COMPLEXED WITH DIAMINOBENZOPHENONE-BETA,GAMMA-IMIDO-GTP | | Descriptor: | 3-AMINOBENZOPHENONE-4-YL-AMINOHYDROXYPHOSPHINYLAMINOPHOSPHONIC ACID-GUANYLATE ESTER, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Ahmadian, M.R, Zor, T, Vogt, D, Kabsch, W, Selinger, Z, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanosine triphosphatase stimulation of oncogenic Ras mutants.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
521P
 
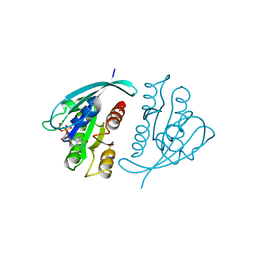 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, H-RAS P21 PROTEIN, MAGNESIUM ION | | Authors: | Schlichting, I, Krengel, U, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
1WQ1
 
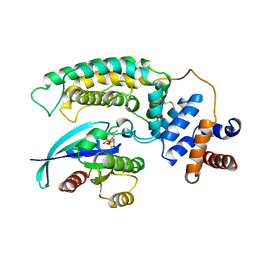 | | RAS-RASGAP COMPLEX | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, H-RAS, ... | | Authors: | Scheffzek, K, Ahmadian, M.R, Kabsch, W, Wiesmueller, L, Lautwein, A, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1997-07-03 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants.
Science, 277, 1997
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | Descriptor: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS6
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | NICKEL (II) ION, PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS4
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BSZ
 
 | | PEPTIDE DEFORMYLASE AS FE2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | FE (III) ION, NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS7
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM | | Descriptor: | NICKEL (II) ION, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1NF1
 
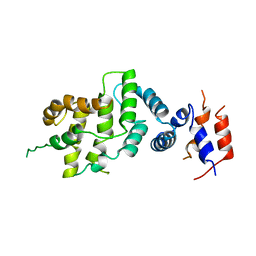 | | THE GAP RELATED DOMAIN OF NEUROFIBROMIN | | Descriptor: | PROTEIN (NEUROFIBROMIN) | | Authors: | Scheffzek, K, Ahmadian, M.R, Wiesmueller, L, Kabsch, W, Stege, P, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1998-07-08 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the GAP-related domain from neurofibromin and its implications.
EMBO J., 17, 1998
|
|
1CM5
 
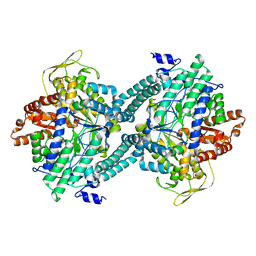 | | CRYSTAL STRUCTURE OF C418A,C419A MUTANT OF PFL FROM E.COLI | | Descriptor: | CARBONATE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-14 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1CRK
 
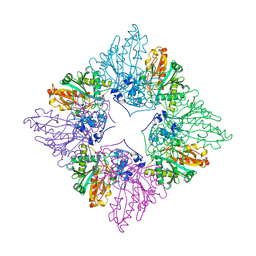 | | MITOCHONDRIAL CREATINE KINASE | | Descriptor: | CREATINE KINASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Schnyder, T, Wallimann, T, Kabsch, W. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of mitochondrial creatine kinase.
Nature, 381, 1996
|
|
121P
 
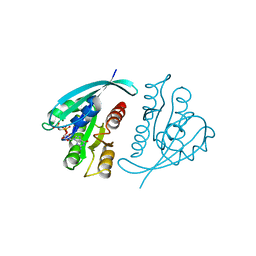 | | STRUKTUR UND GUANOSINTRIPHOSPHAT-HYDROLYSEMECHANISMUS DES C-TERMINAL VERKUERZTEN MENSCHLICHEN KREBSPROTEINS P21-H-RAS | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Krengel, U, Scheffzek, K, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Struktur Und Guanosintriphosphat-Hydrolysemechanismus Des C-Terminal Verkuerzten Menschlichen Krebsproteins P21-H-Ras
Thesis, 1991
|
|
1NDA
 
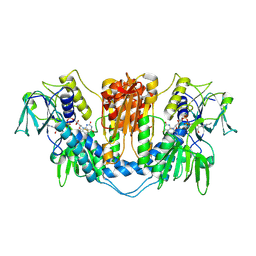 | | THE STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN THE OXIDIZED AND NADPH REDUCED STATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE OXIDOREDUCTASE | | Authors: | Lantwin, C.B, Kabsch, W, Pai, E.F, Schlichting, I, Krauth-Siegel, R.L. | | Deposit date: | 1993-07-02 | | Release date: | 1994-09-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of Trypanosoma cruzi trypanothione reductase in the oxidized and NADPH reduced state.
Proteins, 18, 1994
|
|
1QH4
 
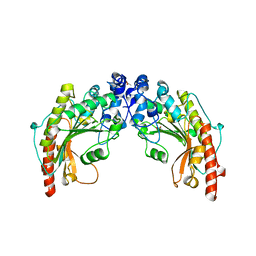 | | CRYSTAL STRUCTURE OF CHICKEN BRAIN-TYPE CREATINE KINASE AT 1.41 ANGSTROM RESOLUTION | | Descriptor: | ACETATE ION, CALCIUM ION, CREATINE KINASE | | Authors: | Eder, M, Schlattner, U, Becker, A, Wallimann, T, Kabsch, W, Fritz-Wolf, K. | | Deposit date: | 1999-05-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of brain-type creatine kinase at 1.41 A resolution.
Protein Sci., 8, 1999
|
|
1QK1
 
 | | CRYSTAL STRUCTURE OF HUMAN UBIQUITOUS MITOCHONDRIAL CREATINE KINASE | | Descriptor: | CREATINE KINASE, UBIQUITOUS MITOCHONDRIAL, PHOSPHATE ION | | Authors: | Eder, M, Schlattner, U, Fritz-Wolf, K, Wallimann, T, Kabsch, W. | | Deposit date: | 1999-07-08 | | Release date: | 2000-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Ubiquitous Mitochondrial Creatine Kinase
Proteins: Struct.,Funct., Genet., 39, 2000
|
|
1RVD
 
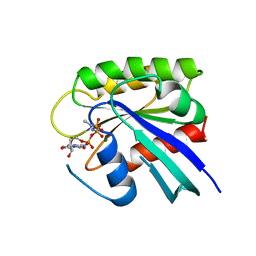 | | H-RAS COMPLEXED WITH DIAMINOBENZOPHENONE-BETA,GAMMA-IMIDO-GTP | | Descriptor: | 3-AMINOBENZOPHENONE-4-YL-AMINOHYDROXYPHOSPHINYLAMINOPHOSPHONIC ACID-GUANYLATE ESTER, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Ahmadian, M.R, Zor, T, Vogt, D, Kabsch, W, Selinger, Z, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Guanosine triphosphatase stimulation of oncogenic Ras mutants.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1WER
 
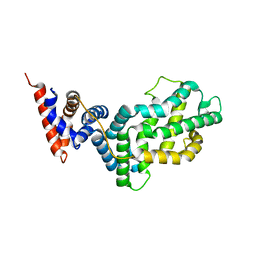 | | RAS-GTPASE-ACTIVATING DOMAIN OF HUMAN P120GAP | | Descriptor: | P120GAP | | Authors: | Scheffzek, K, Lautwein, A, Kabsch, W, Ahmadian, M.R, Wittinghofer, A. | | Deposit date: | 1996-11-20 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras.
Nature, 384, 1996
|
|
1GK0
 
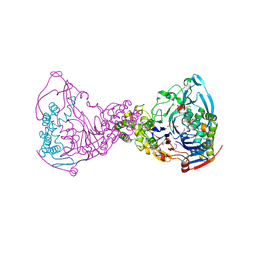 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | 1,2-ETHANEDIOL, CEPHALOSPORIN ACYLASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
1GK1
 
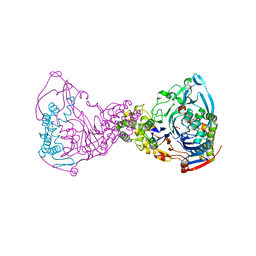 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | CEPHALOSPORIN ACYLASE, GLYCEROL | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
221P
 
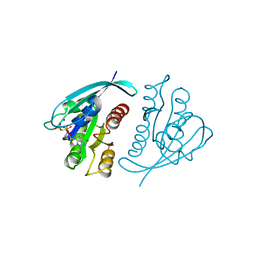 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Krengel, U, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
1ICJ
 
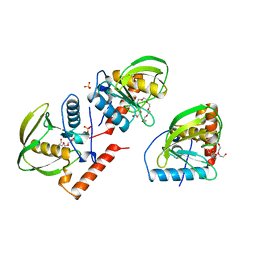 | | PDF PROTEIN IS CRYSTALLIZED AS NI2+ CONTAINING FORM, COCRYSTALLIZED WITH INHIBITOR POLYETHYLENE GLYCOL (PEG) | | Descriptor: | NICKEL (II) ION, NONAETHYLENE GLYCOL, PEPTIDE DEFORMYLASE, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1K8R
 
 | | Crystal structure of Ras-Bry2RBD complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein kinase byr2, ... | | Authors: | Scheffzek, K, Gruenewald, P, Wohlgemuth, S, Kabsch, W, Tu, H, Wigler, M, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2001-10-25 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Ras-Byr2RBD complex: structural basis for Ras effector recognition in yeast.
Structure, 9, 2001
|
|
1G6T
 
 | | STRUCTURE OF EPSP SYNTHASE LIGANDED WITH SHIKIMATE-3-PHOSPHATE | | Descriptor: | EPSP SYNTHASE, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Schonbrunn, E, Eschenburg, S, Shuttleworth, W, Schloss, J.V, Amrhein, N, Evans, J.N.S, Kabsch, W. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
