6YAV
 
 | | Structure of FeSII (Shethna) protein from Azotobacter vinelandii | | Descriptor: | Dimeric (2Fe-2S) protein, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Kabasakal, B.V, Kung, W.K.A, Cotton, C.A.R, Lieber, L, McFarlane, C.R, Murray, J.W. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of FeSII (Shethna) protein from Azotobacter vinelandii
To Be Published
|
|
8AEQ
 
 | |
8AEO
 
 | |
8AET
 
 | |
8A8T
 
 | |
8A7S
 
 | |
8AEW
 
 | |
8AER
 
 | |
8A30
 
 | |
4EOC
 
 | |
4EOE
 
 | |
4EOD
 
 | |
5M45
 
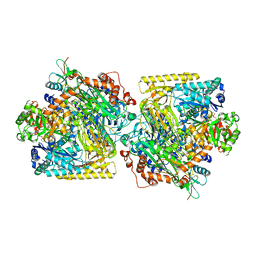 | | Structure of Acetone Carboxylase purified from Xanthobacter autotrophicus | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kabasakal, B.V, Wells, J.N, Nwaobi, B.C, Eilers, B.J, Peters, J.W, Murray, J.W. | | Deposit date: | 2016-10-18 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
5DRA
 
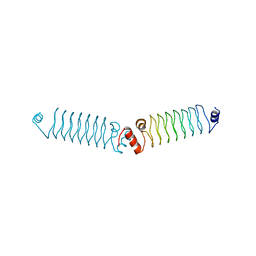 | | t3284 loop variant of beta1 | | Descriptor: | 3beta1 | | Authors: | Kabasakal, B.V, MacDonald, J.T, Murray, W.W. | | Deposit date: | 2015-09-15 | | Release date: | 2016-08-31 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DZB
 
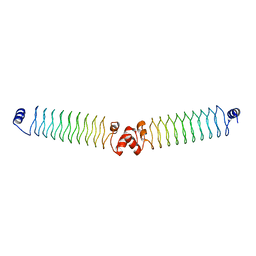 | | 6beta1 | | Descriptor: | 6beta1 | | Authors: | Kabasakal, B.V, MacDonald, J.T, Murray, W.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-08-31 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DQA
 
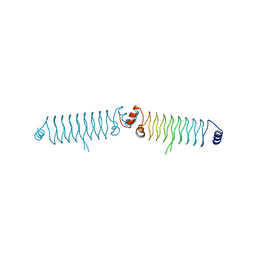 | | t3284 loop variant of beta1 | | Descriptor: | t3284 | | Authors: | Kabasakal, B.V, MacDonald, J.T, Murray, W.W. | | Deposit date: | 2015-09-14 | | Release date: | 2016-08-31 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5SVB
 
 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase AMP bound structure | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
5SVC
 
 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase nucleotide-free structure | | Descriptor: | Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, Acetone carboxylase gamma subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
5DI5
 
 | | beta1 t801 loop variant in P3221 | | Descriptor: | beta1 t801 loop variant | | Authors: | MacDonald, J.T, Kabasakal, B.V, Murray, J.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-08-31 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DNS
 
 | | t1428 loop variant in P3221 | | Descriptor: | t1428 beta1 loop design | | Authors: | MacDonald, J.T, Kabasakal, B.V, Murray, J.W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-28 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (3.561 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DN0
 
 | | t1555 loop variant | | Descriptor: | beta1 t1555 loop variant | | Authors: | MacDonald, J.T, Kabasakal, B.V, Murray, J.W. | | Deposit date: | 2015-09-09 | | Release date: | 2016-08-31 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FRT
 
 | |
6RK0
 
 | | Structure of the Flavocytochrome Anf3 from Azotobacter vinelandii | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Murray, J.W, Varghese, F, Kabasakal, B. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A low-potential terminal oxidase associated with the iron-only nitrogenase from the nitrogen-fixing bacteriumAzotobacter vinelandii.
J.Biol.Chem., 294, 2019
|
|
8H3W
 
 | |
7Y6A
 
 | | Crystal structure of Chicken Egg Lysozyme | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | DeMirci, H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryogenic X-ray crystallographic studies of biomacromolecules at Turkish Light Source " Turkish DeLight ".
Turk J Biol, 47, 2023
|
|
