4UHP
 
 | | Crystal structure of the pyocin AP41 DNase-Immunity complex | | Descriptor: | BACTERIOCIN IMMUNITY PROTEIN, LARGE COMPONENT OF PYOCIN AP41 | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
4UHQ
 
 | | Crystal structure of the pyocin AP41 DNase | | Descriptor: | CITRIC ACID, LARGE COMPONENT OF PYOCIN AP41, NICKEL (II) ION | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
3ZPT
 
 | | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[(4-bromophenyl)methyl]-2-[3-[(3Z,8S,11R)-11-oxidanyl-7,10-bis(oxidanylidene)-8-propan-2-yl-6,9-diazabicyclo[11.2.2]heptadeca-1(16),3,13(17),14-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Joshi, A, Veron, J.B, Unge, J, Rosenquist, A, Wallberg, H, Samuelsson, B, Hallberg, A, Larhed, M. | | Deposit date: | 2013-03-01 | | Release date: | 2013-11-06 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors
J.Med.Chem., 56, 2013
|
|
3ZPS
 
 | | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[(4-bromophenyl)methyl]-2-[(4R)-5-[[(2S)-3,3-dimethyl-1-oxidanylidene-1-(prop-2-enylamino)butan-2-yl]amino]-4-oxidanyl-5-oxidanylidene-4-(phenylmethyl)pentyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Joshi, A, Veron, J.B, Unge, J, Rosenquist, A, Wallberg, H, Samuelsson, B, Hallberg, A, Larhed, M. | | Deposit date: | 2013-03-01 | | Release date: | 2013-11-06 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors.
J.Med.Chem., 56, 2013
|
|
3ZPU
 
 | | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[(4-bromophenyl)methyl]-2-[3-[(3Z,8S,11R)-8-tert-butyl-11-oxidanyl-7,10-bis(oxidanylidene)-6,9-diazabicyclo[11.2.2]heptadeca-1(15),3,13,16-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Joshi, A, Veron, J.B, Unge, J, Rosenquist, A, Wallberg, H, Samuelsson, B, Hallberg, A, Larhed, M. | | Deposit date: | 2013-03-01 | | Release date: | 2013-11-06 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors
J.Med.Chem., 56, 2013
|
|
4Z7H
 
 | |
3ZZZ
 
 | |
3ZZY
 
 | |
4CQ1
 
 | | Crystal structure of the neuronal isoform of PTB | | Descriptor: | CHLORIDE ION, POLYPYRIMIDINE TRACT-BINDING PROTEIN 2, ZINC ION | | Authors: | Joshi, A, Buckroyd, A.N, Curry, S. | | Deposit date: | 2014-02-10 | | Release date: | 2014-02-19 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Solution and Crystal Structures of a C Terminal Fragment of the Neuronal Isoform of the Polypyrimidine Tract Binding Protein (Nptb)
Peerj, 2, 2014
|
|
4BYJ
 
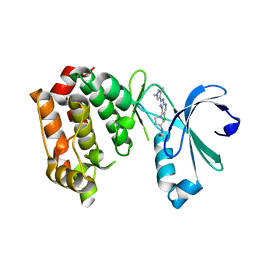 | | Aurora A kinase bound to a highly selective imidazopyridine inhibitor | | Descriptor: | (S)-N-(1-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)pyrrolidin-3-yl)acetamide, AURORA KINASE A | | Authors: | Joshi, A, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Aurora Isoform Selectivity: Design and Synthesis of Imidazo[4,5-B]Pyridine Derivatives as Highly Selective Inhibitors of Aurora-A Kinase in Cells.
J.Med.Chem., 56, 2013
|
|
4BYI
 
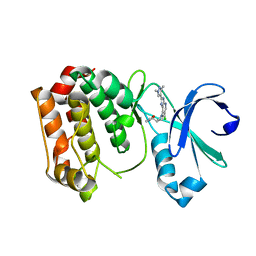 | | Aurora A kinase bound to a highly selective imidazopyridine inhibitor | | Descriptor: | (S)-N-((1-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)pyrrolidin-3-yl)methyl)acetamide, AURORA KINASE A | | Authors: | Joshi, A, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aurora Isoform Selectivity: Design and Synthesis of Imidazo[4,5-B]Pyridine Derivatives as Highly Selective Inhibitors of Aurora-A Kinase in Cells.
J.Med.Chem., 56, 2013
|
|
4Z7G
 
 | |
5ODW
 
 | | Structure of the FpvAI-pyocin S2 complex | | Descriptor: | Ferripyoverdine receptor, Pyocin-S2, SULFATE ION | | Authors: | White, P, Joshi, A, Kleanthous, C. | | Deposit date: | 2017-07-06 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Exploitation of an iron transporter for bacterial protein antibiotic import.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2NV3
 
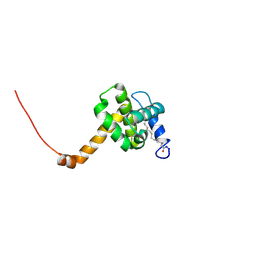 | | Solution structure of L8A mutant of HIV-1 myristoylated matrix protein | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Saad, J.S, Loeliger, E, Luncsford, P, Liriano, M, Tai, J, Kim, A, Miller, J, Joshi, A, Freed, E.O, Summers, M.F. | | Deposit date: | 2006-11-10 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Point mutations in the HIV-1 matrix protein turn off the myristyl switch.
J.Mol.Biol., 366, 2007
|
|
2CG5
 
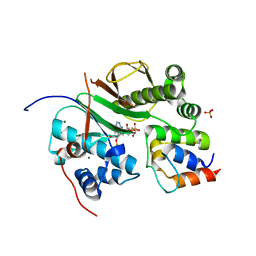 | | Structure of aminoadipate-semialdehyde dehydrogenase- phosphopantetheinyl transferase in complex with cytosolic acyl carrier protein and coenzyme A | | Descriptor: | COENZYME A, FATTY ACID SYNTHASE, L-AMINOADIPATE-SEMIALDEHYDE DEHYDROGENASE-PHOSPHOPANTETHEINYL TRANSFERASE, ... | | Authors: | Bunkoczi, G, Joshi, A, Papagrigoriu, E, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, von Delft, F, Smith, S, Oppermann, U. | | Deposit date: | 2006-02-27 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and Substrate Recognition of Human Holo Acp Synthase.
Chem.Biol., 14, 2007
|
|
2PO8
 
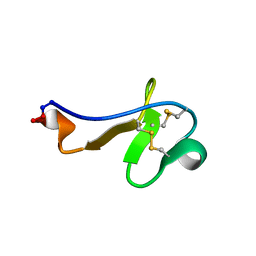 | |
2JMG
 
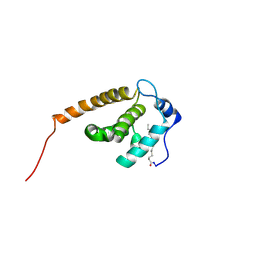 | | Solution structure of V7R mutant of HIV-1 myristoylated matrix protein | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Saad, J.S, Loeliger, E, Luncsford, P, Liriano, M, Tai, J, Kim, A, Miller, J, Joshi, A, Freed, E.O, Summers, M.F. | | Deposit date: | 2006-11-11 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Point mutations in the HIV-1 matrix protein turn off the myristyl switch.
J.Mol.Biol., 366, 2007
|
|
2MJU
 
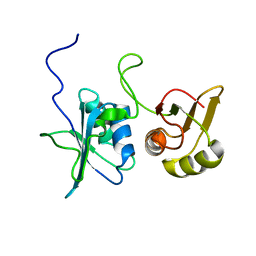 | |
8DYA
 
 | |
7MKM
 
 | |
7MKL
 
 | |
5EW5
 
 | |
6O3T
 
 | | Structural basis of FOXC2 and DNA interactions | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein C2 | | Authors: | Nam, H.-J, Li, S. | | Deposit date: | 2019-02-27 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Crystal Structure of FOXC2 in Complex with DNA Target.
Acs Omega, 4, 2019
|
|
5DH4
 
 | | PDE10 complexed with 5-chloro-N-[(2,4-dimethylthiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, 5-chloro-N-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6HX1
 
 | | IRE1 ALPHA IN COMPLEX WITH imidazo[1,2-b]pyridazin-8-amine compound 2 | | Descriptor: | 6-chloranyl-~{N}-(cyclopropylmethyl)-3-(2~{H}-indazol-5-yl)imidazo[1,2-b]pyridazin-8-amine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Augustin, M.A, Krapp, S, Bayliss, R, Collins, I. | | Deposit date: | 2018-10-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Binding to an Unusual Inactive Kinase Conformation by Highly Selective Inhibitors of Inositol-Requiring Enzyme 1 alpha Kinase-Endoribonuclease.
J.Med.Chem., 62, 2019
|
|
