1QQO
 
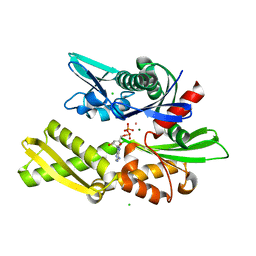 | | E175S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, HYDROLASE (ACTING ON ACID ANHYDRIDES), ... | | Authors: | Johnson, E.R, McKay, D.B. | | Deposit date: | 1999-06-07 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein.
Biochemistry, 38, 1999
|
|
1QQM
 
 | | D199S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, D199S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN, ... | | Authors: | Johnson, E.R, Mckay, D.B. | | Deposit date: | 1999-06-07 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein.
Biochemistry, 38, 1999
|
|
1QQN
 
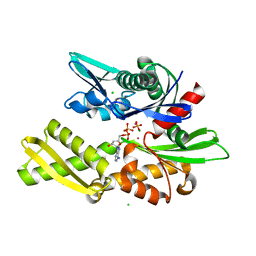 | | D206S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, D206S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN, ... | | Authors: | Johnson, E.R, McKay, D.B. | | Deposit date: | 1999-06-07 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein.
Biochemistry, 38, 1999
|
|
1QVA
 
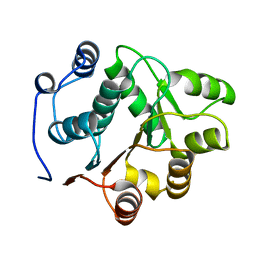 | | YEAST INITIATION FACTOR 4A N-TERMINAL DOMAIN | | Descriptor: | INITIATION FACTOR 4A | | Authors: | Johnson, E.R, McKay, D.B. | | Deposit date: | 1999-07-07 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase
RNA, 5, 1999
|
|
1FUU
 
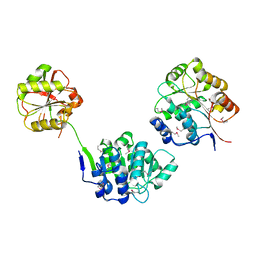 | | YEAST INITIATION FACTOR 4A | | Descriptor: | YEAST INITIATION FACTOR 4A | | Authors: | Caruthers, J.M, Johnson, E.R, McKay, D.B. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FUK
 
 | |
6VIM
 
 | | P. putida mandelate racemase co-crystallized with phenylboronic acid | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mandelate racemase, ... | | Authors: | Grandinetti, L, Sharma, A.N, Bearne, S.L, St Maurice, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent Inhibition of Mandelate Racemase by Boronic Acids: Boron as a Mimic of a Carbon Acid Center.
Biochemistry, 59, 2020
|
|
