2VYA
 
 | | Crystal Structure of fatty acid amide hydrolase conjugated with the drug-like inhibitor PF-750 | | Descriptor: | 4-(quinolin-3-ylmethyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Johnson, D.S, Wang, Z, Everdeen, D.S, Liimatta, M, Pabst, B, Bhattacharya, K, Nugent, R.A, Kamtekar, S, Cravatt, B.F, Ahn, K, Stevens, R.C. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Guided Inhibitor Design for Human Faah by Interspecies Active Site Conversion.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1AG5
 
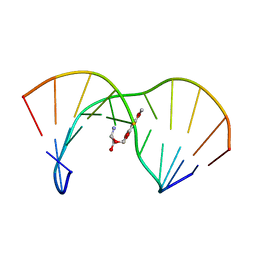 | | THE SOLUTION STRUCTURE OF AN AFLATOXIN B1 EPOXIDE ADDUCT AT THE N7 POSITION OF GUANINE OPPOSITE AN ADENINE IN THE COMPLEMENTARY STRAND OF AN OLIGODEOXYNUCLEOTIDE DUPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1, DNA (5'-D(*CP*CP*AP*TP*CP*GP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*AP*GP*AP*TP*GP*G)-3') | | Authors: | Johnson, D.S, Stone, M.P. | | Deposit date: | 1997-04-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 opposite CpA in the complementary strand of an oligodeoxynucleotide duplex as determined by 1H NMR.
Biochemistry, 34, 1995
|
|
6AX1
 
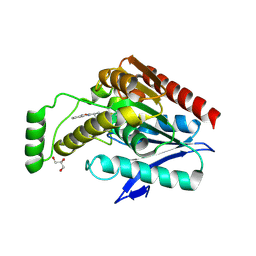 | | Structure of human monoacylglycerol lipase bound to a covalent inhibitor | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-yl 3-(3-phenyl-1,2,4-oxadiazol-5-yl)azetidine-1-carboxylate, GLYCEROL, Monoglyceride lipase | | Authors: | Pandit, J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Azetidine and Piperidine Carbamates as Efficient, Covalent Inhibitors of Monoacylglycerol Lipase.
J. Med. Chem., 60, 2017
|
|
6BQ0
 
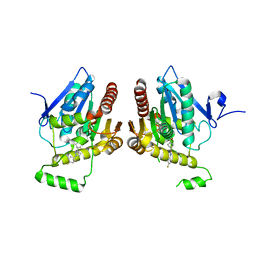 | | Structure of human monoacylglycerol lipase bound to a covalent inhibitor | | Descriptor: | 1-({(1R,5S,6r)-6-[1-(4-fluorophenyl)-1H-pyrazol-3-yl]-3-azabicyclo[3.1.0]hexane-3-carbonyl}oxy)pyrrolidine-2,5-dione, Monoglyceride lipase | | Authors: | Pandit, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Trifluoromethyl Glycol Carbamates as Potent and Selective Covalent Monoacylglycerol Lipase (MAGL) Inhibitors for Treatment of Neuroinflammation.
J. Med. Chem., 61, 2018
|
|
2WAP
 
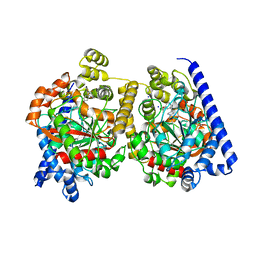 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with the drug-like urea inhibitor PF-3845 | | Descriptor: | 4-(3-{[5-(trifluoromethyl)pyridin-2-yl]oxy}benzyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Kamtekar, S, Stevens, R.C. | | Deposit date: | 2009-02-11 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Highly Selective Faah Inhibitor that Reduces Inflammatory Pain.
Chem.Biol., 16, 2009
|
|
1DSI
 
 | | Solution structure of a duocarmycin sa-indole-alkylated dna dupleX | | Descriptor: | 4-HYDROXY-6-(1H-INDOLE-2-CARBONYL)-8-METHYL-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Schnell, J.R, Ketchem, R.R, Boger, D.L, Chazin, W.J. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Binding-Induced Activation of DNA Alkylation by Duocarmycin SA: Insights from the Structure of an Indole Derivative-DNA Adduct
J.Am.Chem.Soc., 121, 1999
|
|
1E74
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R11E | | Descriptor: | ALPHA-CONOTOXIN IM1(R11E) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E75
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R7L | | Descriptor: | ALPHA-CONOTOXIN IM1(R7L) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E76
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT D5N | | Descriptor: | ALPHA-CONOTOXIN IM1(D5N) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1IM1
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
