1MTX
 
 | | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF MARGATOXIN BY 1H, 13C, 15N TRIPLE-RESONANCE NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | MARGATOXIN | | Authors: | Johnson, B.A, Stevens, S.P, Williamson, J.M. | | Deposit date: | 1994-12-27 | | Release date: | 1995-11-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of margatoxin by 1H, 13C, 15N triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 33, 1994
|
|
1BM6
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN STROMELYSIN-1 COMPLEXED TO A POTENT NON-PEPTIDIC INHIBITOR, NMR, 20 STRUCTURES | | Descriptor: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | Authors: | Li, Y, Zhang, X, Melton, R, Ganu, V, Gonnella, N.C. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin-1 complexed to a potent, nonpeptidic inhibitor.
Biochemistry, 37, 1998
|
|
3HEF
 
 | |
8TH5
 
 | |
8TH1
 
 | |
8TH7
 
 | |
8TH6
 
 | |
2SRT
 
 | |
2N1Q
 
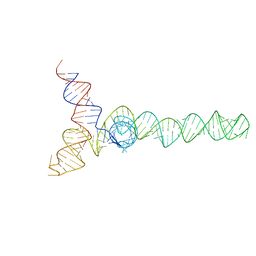 | | HIV-1 Core Packaging Signal | | Descriptor: | RNA_(155-MER) | | Authors: | Keane, S.C, Summers, M.F. | | Deposit date: | 2015-04-15 | | Release date: | 2015-05-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | RNA structure. Structure of the HIV-1 RNA packaging signal.
Science, 348, 2015
|
|
1HFS
 
 | |
1SLM
 
 | |
1SLN
 
 | |
