4AJG
 
 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, MAGNESIUM ION, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
4AJD
 
 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | 2-ETHYL-4-METHYL-PHTHALAZIN-1-ONE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
6EJ2
 
 | | BACE1 compound 28 | | Descriptor: | Beta-secretase 1, compound 28 | | Authors: | Johansson, P. | | Deposit date: | 2017-09-20 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Toward beta-Secretase-1 Inhibitors with Improved Isoform Selectivity.
J. Med. Chem., 61, 2018
|
|
6EJ3
 
 | | BACE1 compound 23 | | Descriptor: | (1r,4r)-4-methoxy-6'-(5-methyl-3-pyridinyl)-3'H-dispiro[cyclohexane-1,2'-indene-1',4''-[1,3]oxazol]-2''-amine, Beta-secretase 1 | | Authors: | Johansson, P. | | Deposit date: | 2017-09-20 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Toward beta-Secretase-1 Inhibitors with Improved Isoform Selectivity.
J. Med. Chem., 61, 2018
|
|
4AJF
 
 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, MAGNESIUM ION, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
3HMJ
 
 | | Saccharomyces cerevisiae FAS type I | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Johansson, P, Mulinacci, B, Koestler, C, Vollrath, R, Oesterhelt, D, Grininger, M. | | Deposit date: | 2009-05-29 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Multimeric options for the auto-activation of the Saccharomyces cerevisiae FAS type I megasynthase
Structure, 17, 2009
|
|
2VKZ
 
 | | Structure of the cerulenin-inhibited fungal fatty acid synthase type I multienzyme complex | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FATTY ACID SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Johansson, P, Wiltschi, B, Kumari, P, Kessler, B, Vonrhein, C, Vonck, J, Oesterhelt, D, Grininger, M. | | Deposit date: | 2008-01-07 | | Release date: | 2008-08-12 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Inhibition of the Fungal Fatty Acid Synthase Type I Multienzyme Complex.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1UMZ
 
 | | Xyloglucan endotransglycosylase in complex with the xyloglucan nonasaccharide XLLG. | | Descriptor: | XYLOGLUCAN ENDOTRANSGLYCOSYLASE, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[beta-D-galactopyranose-(1-2)-alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A.M, Henriksson, H, Denman, S.E, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
1UN1
 
 | | Xyloglucan endotransglycosylase native structure. | | Descriptor: | GOLD ION, XYLOGLUCAN ENDOTRANSGLYCOSYLASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A, Henriksson, H, Denman, S, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
2WAT
 
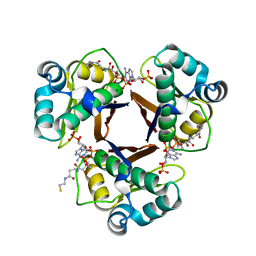 | | Structure of the fungal type I FAS PPT domain in complex with CoA | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Johansson, P, Mulincacci, B, Koestler, C, Grininger, M. | | Deposit date: | 2009-02-15 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multimeric Options for the Auto-Activation of the Saccharomyces Cerevisiae Fas Type I Megasynthase.
Structure, 17, 2009
|
|
2WAS
 
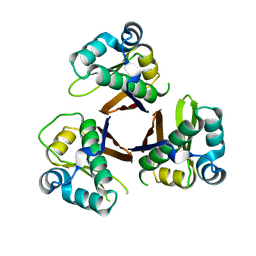 | |
2YOP
 
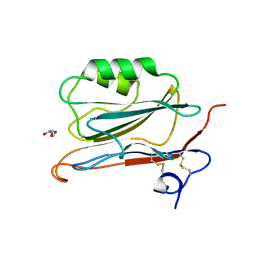 | | Long wavelength S-SAD structure of FAM3B PANDER | | Descriptor: | GLYCEROL, PROTEIN FAM3B | | Authors: | Johansson, P, Bernstrom, J, Gorman, T, Oster, L, Backstrom, S, Schweikart, F, Xu, B, Xue, Y, Holmberg Schiavone, L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fam3B Pander and Fam3C Ilei Represent a Distinct Class of Signaling Molecules with a Non-Cytokine-Like Fold.
Structure, 21, 2013
|
|
2YOQ
 
 | | Structure of FAM3B PANDER E30 construct | | Descriptor: | GLYCEROL, PROTEIN FAM3B | | Authors: | Johansson, P, Bernstrom, J, Gorman, T, Oster, L, Backstrom, S, Schweikart, F, Xu, B, Xue, Y, Holmberg Schiavone, L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fam3B Pander and Fam3C Ilei Represent a Distinct Class of Signaling Molecules with a Non-Cytokine-Like Fold.
Structure, 21, 2013
|
|
2BNG
 
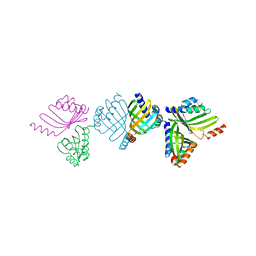 | | Structure of an M.tuberculosis LEH-like epoxide hydrolase | | Descriptor: | CALCIUM ION, MB2760 | | Authors: | Johansson, P, Arand, M, Unge, T, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2005-03-24 | | Release date: | 2005-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an Atypical Epoxide Hydrolase from Mycobacterium Tuberculosis Gives Insights Into its Function.
J.Mol.Biol., 351, 2005
|
|
2C2I
 
 | | Structure and function of Rv0130, a conserved hypothetical protein from M.tuberculosis | | Descriptor: | RV0130 | | Authors: | Johansson, P, Castell, A, Jones, T.A, Backbro, K. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of Rv0130, a Conserved Hypothetical Protein from Mycobacterium Tuberculosis.
Protein Sci., 15, 2006
|
|
7Q6S
 
 | | Keap1 compound complex | | Descriptor: | (5S,8R)-16-(2,1,3-benzoxadiazol-4-yl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q6Q
 
 | | Keap1 compound complex | | Descriptor: | (5S,8R)-N,N-dimethyl-8-[[(2S)-1-[4-(methylamino)-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(12),13,15-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Johansson, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q96
 
 | | Keap1 compound complex | | Descriptor: | 4-[(5S,8R)-5-(dimethylcarbamoyl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-trien-16-yl]benzoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q8R
 
 | | Keap1 compound complex | | Descriptor: | 3-[(5S,8R)-5-(dimethylcarbamoyl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-trien-16-yl]benzoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q5H
 
 | | Keap1 compound complex | | Descriptor: | (3S,5S,8R)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-3,7,11-tris(oxidanylidene)-10-oxa-3$l^{4}-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
5JNQ
 
 | | MraY tunicamycin complex | | Descriptor: | PALMITIC ACID, Phospho-N-acetylmuramoyl-pentapeptide-transferase, Tunicamycin, ... | | Authors: | Johansson, P. | | Deposit date: | 2016-04-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MraY-antibiotic complex reveals details of tunicamycin mode of action.
Nat. Chem. Biol., 13, 2017
|
|
6Z6A
 
 | | Keap1 macrocycle complex | | Descriptor: | (5S,8R)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Johansson, P. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mining Natural Products for Macrocycles to Drug Difficult Targets.
J.Med.Chem., 64, 2021
|
|
5LC3
 
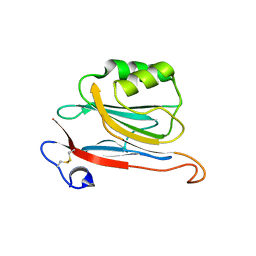 | | Xray structure of mouse FAM3C ILEI monomer | | Descriptor: | Protein FAM3C | | Authors: | Johansson, P, Jansson, A. | | Deposit date: | 2016-06-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer.
J. Biol. Chem., 292, 2017
|
|
5LC2
 
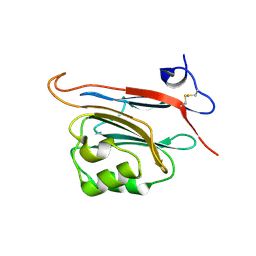 | | Xray structure of human FAM3C ILEI monomer | | Descriptor: | Protein FAM3C | | Authors: | Johansson, P, Jansson, A. | | Deposit date: | 2016-06-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer.
J. Biol. Chem., 292, 2017
|
|
5LC4
 
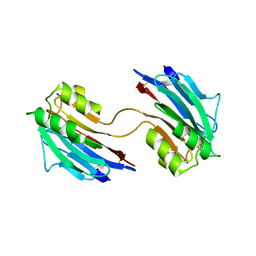 | | Xray structure of mouse FAM3C ILEI dimer | | Descriptor: | Protein FAM3C | | Authors: | Johansson, P, Jansson, A. | | Deposit date: | 2016-06-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer.
J. Biol. Chem., 292, 2017
|
|
