4ID7
 
 | | ACK1 kinase in complex with the inhibitor cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Descriptor: | Activated CDC42 kinase 1, SULFATE ION, cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Authors: | Jin, M, Wang, J, Kleinberg, A, Kadalbajoo, M, Siu, K, Cooke, A, Bittner, M, Yao, Y, Thelemann, A, Ji, Q, Bhagwat, S, Crew, A.P, Pachter, J, Epstein, D, Mulvihill, M.J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, selective and orally bioavailable imidazo[1,5-a]pyrazine derived ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6KRE
 
 | | TRiC at 0.05 mM ADP-AlFx, Conformation 2, 0.05-C2 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KRD
 
 | | TRiC at 0.05 mM ADP-AlFx, Conformation 4, 0.05-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS8
 
 | | TRiC at 0.1 mM ADP-AlFx, Conformation 4, 0.1-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS7
 
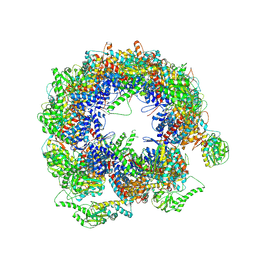 | | TRiC at 0.1 mM ADP-AlFx, Conformation 1, 0.1-C1 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.62 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS6
 
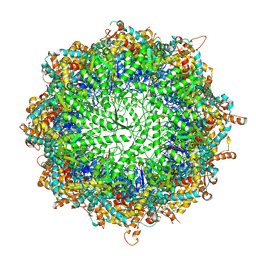 | | TRiC at 0.2 mM ADP-AlFx, Conformation 1, 0.2-C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5H1L
 
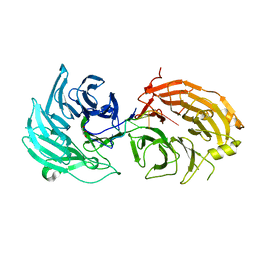 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 7-nt U4 snRNA fragment | | Descriptor: | GLYCEROL, Gem-associated protein 5, U4 snRNA (5'-R(*AP*UP*UP*UP*UP*UP*G)-3') | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5H1M
 
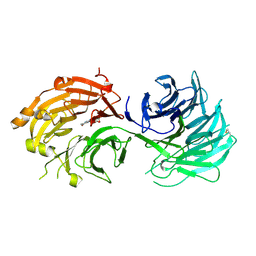 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with M7G | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Gem-associated protein 5 | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5H1K
 
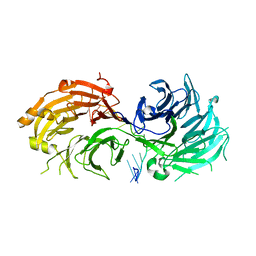 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 13-nt U4 snRNA fragment | | Descriptor: | Gem-associated protein 5, U4 snRNA (5'-R(*GP*CP*AP*AP*UP*UP*UP*UP*UP*GP*AP*CP*A)-3') | | Authors: | Wang, Y, Jin, W, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5H1J
 
 | | Crystal structure of WD40 repeat domains of Gemin5 | | Descriptor: | Gem-associated protein 5 | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
6PWC
 
 | | A complex structure of arrestin-2 bound to neurotensin receptor 1 | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Yin, W, Li, Z, Jin, M, Yin, Y.-L, de Waal, P.W, Pal, K, Gao, X, He, Y, Gao, J, Wang, X, Zhang, Y, Zhou, H, Melcher, K, Jiang, Y, Cong, Y, Zhou, X.E, Yu, X, Xu, H.E. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | A complex structure of arrestin-2 bound to a G protein-coupled receptor.
Cell Res., 29, 2019
|
|
4KNB
 
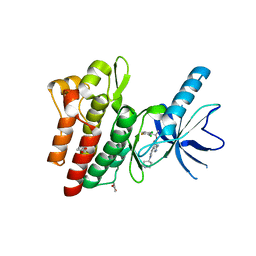 | | C-Met in complex with OSI ligand | | Descriptor: | 7-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[3,2-c]pyridin-6-amine, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor | | Authors: | Wang, J, Steinig, A.G, Li, A.H, Chen, X, Dong, H, Ferraro, C, Jin, M, Kadalbajoo, M, Kleinberg, A, Stolz, K.M, Tavares-Greco, P.A, Wang, T, Albertella, M.R, Peng, Y, Crew, L, Kahler, J. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel 6-aminofuro[3,2-c]pyridines as potent, orally efficacious inhibitors of cMET and RON kinases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8X6M
 
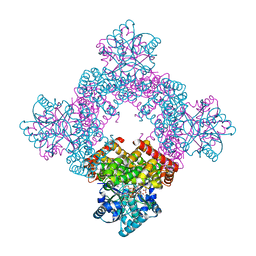 | | Crystal Structure of Glycerol Dehydrogenase in the Presence of NAD+ and Glycerol | | Descriptor: | GLYCEROL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Kang, J.Y, Jin, M, Yang, J, Kim, H, Noh, C, Eom, S.H. | | Deposit date: | 2023-11-21 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the octamerization of glycerol dehydrogenase.
Plos One, 19, 2024
|
|
5JDB
 
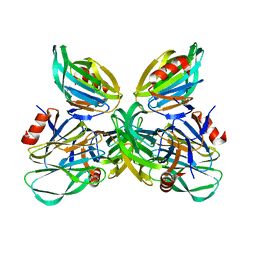 | | Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens | | Descriptor: | Outer capsid protein VP4 | | Authors: | Sun, X, Guo, N, Li, D, Zhou, Y, Jin, M, Xie, G, Pang, L, Zhang, Q, Cao, Y, Duan, Z. | | Deposit date: | 2016-04-16 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens.
Virology, 495, 2016
|
|
5GW4
 
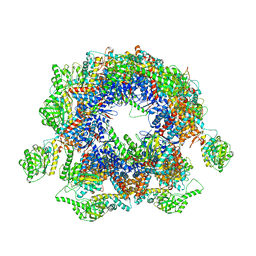 | | Structure of Yeast NPP-TRiC | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GW5
 
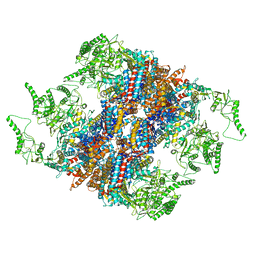 | | Structure of TRiC-AMP-PNP | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
4IP3
 
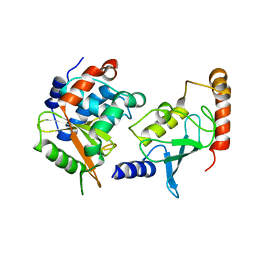 | | Complex structure of OspI and Ubc13 | | Descriptor: | ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Fu, P, Jin, M, Zhang, X, Xu, L, Xia, Z, Zhu, Y. | | Deposit date: | 2013-01-09 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Analysis of Ubc13 Inactivation
To be Published
|
|
5WCO
 
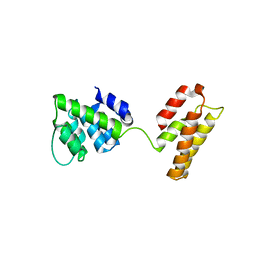 | | Matrix Protein (M1) of Infectious Salmon Anaemia Virus | | Descriptor: | NS2 | | Authors: | Zhang, W, Zheng, W, Toh, Y, Betancourt, M.A, Tu, J, Fan, Y, Vakharia, V, Liu, J, McNew, J.A, Jin, M, Tao, Y.J. | | Deposit date: | 2017-07-01 | | Release date: | 2017-08-16 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of an orthomyxovirus matrix protein reveals mechanisms for self-polymerization and membrane association.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7CLT
 
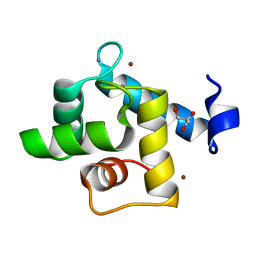 | | Crystal structure of the EFhd1/Swiprosin-2, a mitochondrial actin-binding protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Park, K.R, Lee, Y, Kang, J.Y, Park, T, Jin, M, Yang, J, Jun, C.D, Eom, S.H. | | Deposit date: | 2020-07-22 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07380986 Å) | | Cite: | Structural and Biochemical Characterization of EFhd1/Swiprosin-2, an Actin-Binding Protein in Mitochondria.
Front Cell Dev Biol, 8, 2020
|
|
8GOB
 
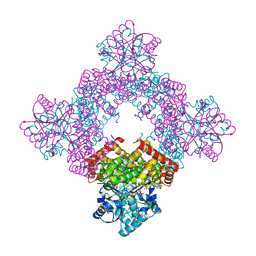 | | Crystal Structure of Glycerol Dehydrogenase in the presence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
8GOA
 
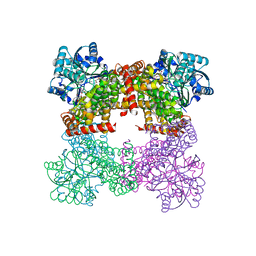 | | Crystal Structure of Glycerol Dehydrogenase in the absence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, ZINC ION | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
6LE5
 
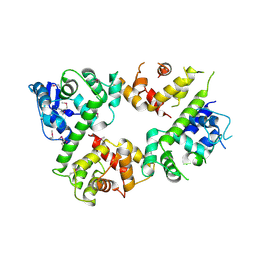 | | Crystal structure of the mitochondrial calcium uptake 1 and 2 heterodimer (MICU1-MICU2 heterodimer) in an apo state | | Descriptor: | Calcium uptake protein 1, mitochondrial, Calcium uptake protein 2 | | Authors: | Park, J, Lee, Y, Park, T, Kang, J.Y, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2019-11-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the MICU1-MICU2 heterodimer provides insights into the gatekeeping threshold shift.
Iucrj, 7, 2020
|
|
