2MO0
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, TaCsp | | Descriptor: | Cold-shock DNA-binding domain protein | | Authors: | Jin, B, Jeong, K.W, Kim, Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and flexibility of the thermophilic cold-shock protein of Thermus aquaticus.
Biochem.Biophys.Res.Commun., 451, 2014
|
|
2MO1
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, TaCsp with dT7 | | Descriptor: | Cold-shock DNA-binding domain protein | | Authors: | Jin, B, Jeong, K.W, Kim, Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and flexibility of the thermophilic cold-shock protein of Thermus aquaticus.
Biochem.Biophys.Res.Commun., 451, 2014
|
|
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
7EQ2
 
 | | Crystal structure of GDP-bound Rab1a-T75D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Cao, Y.L, Gu, D.D, Gao, S. | | Deposit date: | 2021-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55090284 Å) | | Cite: | Aurora kinase A-mediated phosphorylation triggers structural alteration of Rab1A to enhance ER complexity during mitosis
Nat.Struct.Mol.Biol., 2024
|
|
8UGC
 
 | |
2VJ2
 
 | | Human Jagged-1, domains DSL and EGFs1-3 | | Descriptor: | D-MALATE, JAGGED-1 | | Authors: | Johnson, S, Cordle, J, Tay, J.Z, Roversi, P, Handford, P.A, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conserved Face of the Jagged/Serrate Dsl Domain is Involved in Notch Trans-Activation and Cis-Inhibition.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VJ3
 
 | | Human Notch-1 EGFs 11-13 | | Descriptor: | CALCIUM ION, CHLORIDE ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1, ... | | Authors: | Johnson, S, Cordle, J, Tay, J.Z, Roversi, P, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Conserved Face of the Jagged/Serrate Dsl Domain is Involved in Notch Trans-Activation and Cis-Inhibition.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3PP1
 
 | | Crystal Structure of the Human Mitogen-activated protein kinase kinase 1 (MEK 1) in complex with ligand and MgATP | | Descriptor: | 3-[(2R)-2,3-dihydroxypropyl]-6-fluoro-5-[(2-fluoro-4-iodophenyl)amino]-8-methylpyrido[2,3-d]pyrimidine-4,7(3H,8H)-dione, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dougan, D.R. | | Deposit date: | 2010-11-23 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of TAK-733, a potent and selective MEK allosteric site inhibitor for the treatment of cancer.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5XV9
 
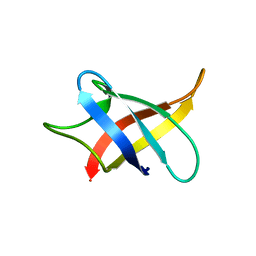 | |
2LXK
 
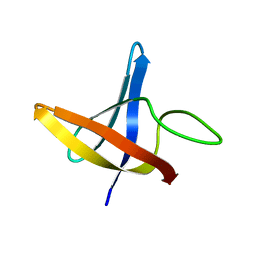 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp | | Descriptor: | Cold shock-like protein CspLA | | Authors: | Lee, J, Jeong, K, Kim, Y. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
2LXJ
 
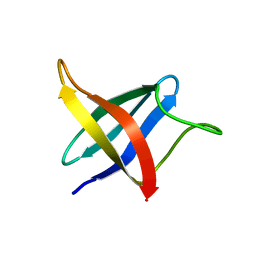 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp with dT7 | | Descriptor: | Cold shock-like protein CspLA | | Authors: | Lee, J, Jeong, K, Kim, Y. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
1NNC
 
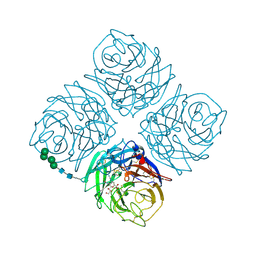 | |
7VPU
 
 | |
7VPS
 
 | |
7VPR
 
 | |
7VPT
 
 | |
7XB5
 
 | |
