1BTL
 
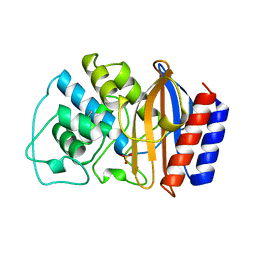 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI TEM1 BETA-LACTAMASE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | BETA-LACTAMASE TEM1, SULFATE ION | | Authors: | Jelsch, C, Mourey, L, Masson, J.M, Samama, J.P. | | Deposit date: | 1993-11-01 | | Release date: | 1995-01-26 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution.
Proteins, 16, 1993
|
|
1EJG
 
 | | CRAMBIN AT ULTRA-HIGH RESOLUTION: VALENCE ELECTRON DENSITY. | | Descriptor: | CRAMBIN (PRO22,SER22/LEU25,ILE25) | | Authors: | Jelsch, C, Teeter, M.M, Lamzin, V, Pichon-Lesme, V, Blessing, B, Lecomte, C. | | Deposit date: | 2000-03-02 | | Release date: | 2000-04-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (0.54 Å) | | Cite: | Accurate protein crystallography at ultra-high resolution: valence electron distribution in crambin.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8PFE
 
 | |
5C7G
 
 | |
1MC2
 
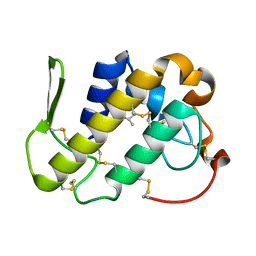 | | monomeric LYS-49 phospholipase A2 homologue purified from AG | | Descriptor: | Acutohaemonlysin, ISOPROPYL ALCOHOL | | Authors: | Liu, Q, Huang, Q.Q, Zhang, R.G, Weeks, C.M, Jelsch, C, Teng, M.K, Niu, L.W. | | Deposit date: | 2002-08-05 | | Release date: | 2002-08-21 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The crystal structure of a novel, inactive, lysine 49 PLA2 from Agkistrodon acutus venom: an ultrahigh resolution, AB initio structure determination
J.Biol.Chem., 278, 2003
|
|
3O4P
 
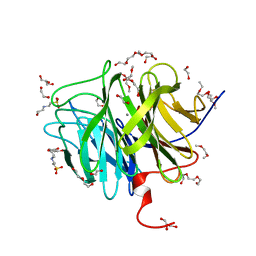 | | DFPase at 0.85 Angstrom resolution (H atoms included) | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Liebschner, D, Elias, M, Koepke, J, Lecomte, C, Guillot, B, Jelsch, C, Chabriere, E. | | Deposit date: | 2010-07-27 | | Release date: | 2011-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Hydrogen atoms in protein structures: high-resolution X-ray diffraction structure of the DFPase.
BMC Res Notes, 6, 2013
|
|
4F1U
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with phosphate at pH 4.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROGENPHOSPHATE ION, Putative alkaline phosphatase, ... | | Authors: | Liebschner, D, Elias, M, Tawfik, D.S, Moniot, S, Fournier, B, Scott, K, Jelsch, C, Guillot, B, Lecomte, C, Chabriere, E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
4F1V
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with phosphate at pH 8.5 | | Descriptor: | HYDROGENPHOSPHATE ION, Putative alkaline phosphatase, SULFATE ION | | Authors: | Liebschner, D, Elias, M, Tawfik, D.S, Moniot, S, Fournier, B, Scott, K, Jelsch, C, Guillot, B, Lecomte, C, Chabriere, E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
1PQ8
 
 | | Trypsin at pH 4 at atomic resolution | | Descriptor: | CITRIC ACID, GLY-GLY-ARG PEPTIDE, LYSINE, ... | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1PQ7
 
 | | Trypsin at 0.8 A, pH5 / borax | | Descriptor: | ARGININE, SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1PQA
 
 | | Trypsin with PMSF at atomic resolution | | Descriptor: | SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1PPZ
 
 | | Trypsin complexes at atomic and ultra-high resolution | | Descriptor: | SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-17 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1PQ5
 
 | | Trypsin at pH 5, 0.85 A | | Descriptor: | ARGININE, SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1BT5
 
 | | CRYSTAL STRUCTURE OF THE IMIPENEM INHIBITED TEM-1 BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PROTEIN (BETA-LACTAMASE), SULFATE ION | | Authors: | Maveyraud, L, Mourey, L, Pedelacq, J.D, Guillet, V, Kotra, L.K, Mobashery, S, Samama, J.P. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Clinical Longevity of Carbapenem Antibiotics in the Face of Challenge by the Common Class A Beta-Lactamases from Antibiotic-Resistant Bacteria
J.Am.Chem.Soc., 120, 1998
|
|
4REK
 
 | | Crystal structure and charge density studies of cholesterol oxidase from Brevibacterium sterolicum at 0.74 ultra-high resolution | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Zarychta, B, Lyubimov, A, Ahmed, M, Munshi, P, Guillot, B, Vrielink, A. | | Deposit date: | 2014-09-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Cholesterol oxidase: ultrahigh-resolution crystal structure and multipolar atom model-based analysis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3NIR
 
 | | Crystal structure of small protein crambin at 0.48 A resolution | | Descriptor: | Crambin, ETHANOL | | Authors: | Schmidt, A, Teeter, M, Weckert, E, Lamzin, V.S. | | Deposit date: | 2010-06-16 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.48 Å) | | Cite: | Crystal structure of small protein crambin at 0.48 A resolution
Acta Crystallogr.,Sect.F, 67, 2011
|
|
