1K85
 
 | | Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 Chitinase A1. | | Descriptor: | CHITINASE A1 | | Authors: | Jee, J.G, Ikegami, T, Hashimoto, M, Kawabata, T, Ikeguchi, M, Watanabe, T, Shirakawa, M. | | Deposit date: | 2001-10-23 | | Release date: | 2002-12-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Fibronectin Type III Domain
from Bacillus circulans WL-12 Chitinase A1
J.Biol.Chem., 277, 2002
|
|
2RQQ
 
 | | Structure of C-terminal region of Cdt1 | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Jee, J.G, Mizuno, T, Kamada, K, Tochio, H, Hiroaki, H, Hanaoka, F, Shirakawa, M. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and mutagenesis studies of the C-terminal region of licensing factor Cdt1 enable the identification of key residues for binding to replicative helicase Mcm proteins
J.Biol.Chem., 285, 2010
|
|
2D07
 
 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
2D2P
 
 | | The solution structure of micelle-bound peptide | | Descriptor: | Pituitary adenylate cyclase activating polypeptide-38 | | Authors: | Tateishi, Y, Jee, J.G, Inooka, H, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-09-14 | | Release date: | 2006-09-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of micelle-bound peptide
To be Published
|
|
1UEL
 
 | | Solution structure of ubiquitin-like domain of hHR23B complexed with ubiquitin-interacting motif of proteasome subunit S5a | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, UV excision repair protein RAD23 homolog B | | Authors: | Fujiwara, K, Tenno, T, Jee, J.G, Sugasawa, K, Ohki, I, Kojima, C, Tochio, H, Hiroaki, H, Hanaoka, H, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-19 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ubiquitin-interacting Motif of S5a Bound to the Ubiquitin-like Domain of HR23B
J.Biol.Chem., 279, 2004
|
|
1UGL
 
 | | Solution structure of S8-SP11 | | Descriptor: | S-locus pollen protein | | Authors: | Mishima, M, Takayama, S, Sasaki, K, Jee, J.G, Kojima, C, Isogai, A, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Male Determinant Factor for Brassica Self-incompatibility
J.Biol.Chem., 278, 2003
|
|
1WYW
 
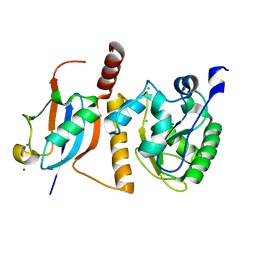 | | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | | Descriptor: | CHLORIDE ION, G/T mismatch-specific thymine DNA glycosylase, MAGNESIUM ION, ... | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-02-17 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymine DNA glycosylase conjugated to SUMO-1.
Nature, 435, 2005
|
|
1WR0
 
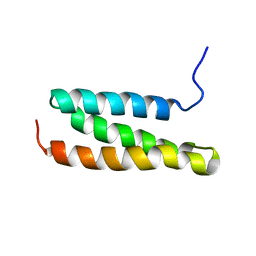 | | Structural characterization of the MIT domain from human Vps4b | | Descriptor: | SKD1 protein | | Authors: | Takasu, H, Jee, J.G, Ohno, A, Goda, N, Fujiwara, K, Tochio, H, Shirakawa, M, Hiroaki, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-07 | | Release date: | 2005-08-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the MIT domain from human Vps4b
Biochem.Biophys.Res.Commun., 334, 2005
|
|
2RRD
 
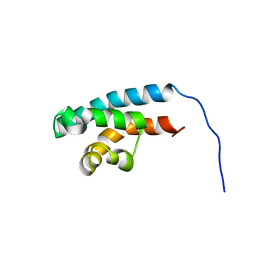 | | Structure of HRDC domain from human Bloom syndrome protein, BLM | | Descriptor: | HRDC domain from Bloom syndrome protein | | Authors: | Sato, A, Mishima, M, Nagai, A, Kim, S.Y, Ito, Y, Hakoshima, T, Jee, J.G, Kitano, K. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HRDC domain of human Bloom syndrome protein BLM
J.Biochem., 148, 2010
|
|
2RRR
 
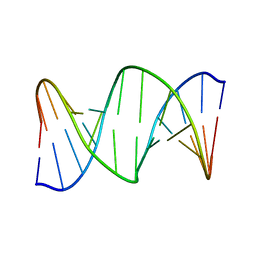 | | DNA oligomer containing ethylene cross-linked cyclic 2'-deoxyuridylate dimer | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*CP*AP*TP*TP*AP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*TP*AP*AP*TP*GP*AP*AP*GP*G)-3') | | Authors: | Furuita, K, Murata, S, Jee, J.G, Ichikawa, S, Matsuda, A, Kojima, C. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural Feature of Bent DNA Recognized by HMGB1
J.Am.Chem.Soc., 133, 2011
|
|
2RRQ
 
 | | DNA oligomer containing propylene cross-linked cyclic 2'-deoxyuridylate dimer | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*CP*AP*(JDT)P*TP*AP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*TP*AP*AP*TP*GP*AP*AP*GP*G)-3') | | Authors: | Furuita, K, Murata, S, Jee, J.G, Ichikawa, S, Matsuda, A, Kojima, C. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Feature of Bent DNA Recognized by HMGB1
J.Am.Chem.Soc., 133, 2011
|
|
7CHT
 
 | | Crystal structure of TTK kinase domain in complex with compound 30 | | Descriptor: | 2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-4-(oxan-4-ylamino)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK, MAGNESIUM ION | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Ko, E.H, Choi, H.G, Son, J.B, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CHM
 
 | | Crystal structure of TTK kinase domain in complex with compound 8 | | Descriptor: | 4-(cyclohexylamino)-2-[(2-methoxy-4-morpholin-4-ylcarbonyl-phenyl)amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CHN
 
 | | Crystal structure of TTK kinase domain in complex with compound 9 | | Descriptor: | 4-(cyclohexylamino)-2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CLH
 
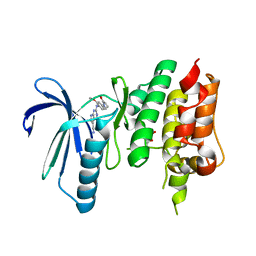 | | Crystal structure of TTK kinase domain in complex with compound 19 | | Descriptor: | 2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-4-(methylamino)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CJA
 
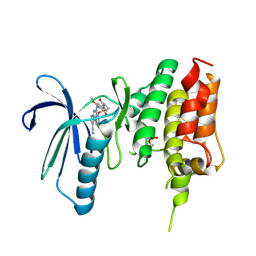 | | Crystal structure of TTK kinase domain in complex with compound 28 | | Descriptor: | 4-(cyclopentylmethylamino)-2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-09 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CIL
 
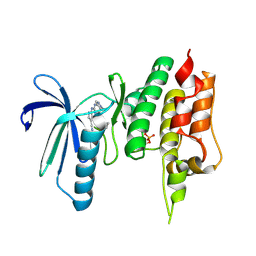 | | Crystal structure of TTK kinase domain in complex with compound 7 | | Descriptor: | 4-(cyclohexylamino)-2-[(1-methylpyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
2JZJ
 
 | | Structure of CrCVNH (C. richardii CVNH) | | Descriptor: | Cyanovirin-N homolog | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2008-01-09 | | Release date: | 2008-03-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
2JZL
 
 | | Structure of NcCVNH (N. CRASSA CVNH) | | Descriptor: | Cyanovirin-N homolog | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2008-01-09 | | Release date: | 2008-03-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
2JZK
 
 | | Structure of TbCVNH (T. BORCHII CVNH) | | Descriptor: | Cyanovirin-N homolog | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2008-01-09 | | Release date: | 2008-03-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
1WR1
 
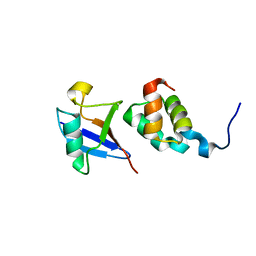 | | The complex structure of Dsk2p UBA with ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin-like protein DSK2 | | Authors: | Ohno, A, Jee, J.G, Fujiwara, K, Tenno, T, Goda, N, Tochio, H, Hiroaki, H, kobayashi, H, Shirakawa, M. | | Deposit date: | 2004-10-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition.
Structure, 13, 2005
|
|
7ER7
 
 | | Crystal structure of hyman Biliverdin IX-beta reductase B with Tamibarotene (A80) | | Descriptor: | 4-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbamoyl]benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ERC
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Deferasirox (ICL) | | Descriptor: | 4-[3,5-bis(2-hydroxyphenyl)-1,2,4-triazol-1-yl]benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ER8
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Sulfasalazine (SAS) | | Descriptor: | 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ER9
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Febuxostat (TEI) | | Descriptor: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
