5DV7
 
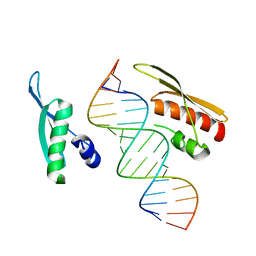 | | Crystal Structure of NF90 tandem dsRBDs with dsRNA | | Descriptor: | Interleukin enhancer-binding factor 3, RNA (5'-R(*CP*CP*AP*GP*CP*AP*UP*UP*AP*UP*GP*AP*AP*AP*GP*UP*GP*A)-3'), RNA (5'-R(*UP*CP*AP*CP*UP*UP*UP*CP*AP*UP*AP*AP*UP*GP*CP*UP*GP*G)-3') | | Authors: | Jayachandran, U, Grey, H, Cook, A.G. | | Deposit date: | 2015-09-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nuclear factor 90 uses an ADAR2-like binding mode to recognize specific bases in dsRNA.
Nucleic Acids Res., 44, 2016
|
|
7AM1
 
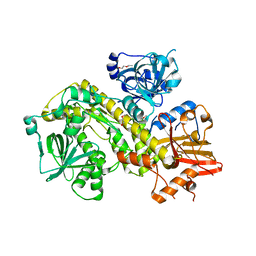 | | Structure of yeast Ssd1, a pseudonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, Protein SSD1 | | Authors: | Cook, A.G, Jayachandran, U. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Yeast Ssd1 is a non-enzymatic member of the RNase II family with an alternative RNA recognition site.
Nucleic Acids Res., 50, 2022
|
|
8A4I
 
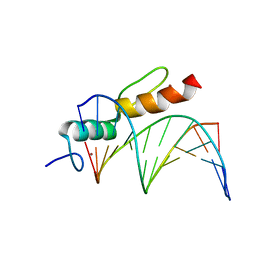 | | Crystal structure of SALL4 zinc finger cluster 4 with AT-rich DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*AP*TP*TP*AP*AP*TP*AP*TP*C)-3'), MAGNESIUM ION, Sal-like protein 4, ... | | Authors: | Watson, J.A, Pantier, R, Jayachandran, U, Chhatbar, K, Alexander-Howden, B, Kruusvee, V, Prendecki, M, Bird, A, Cook, A.G. | | Deposit date: | 2022-06-11 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure of SALL4 zinc finger domain reveals link between AT-rich DNA binding and Okihiro syndrome.
Life Sci Alliance, 6, 2023
|
|
5IW7
 
 | |
4AJ5
 
 | | Crystal structure of the Ska core complex | | Descriptor: | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1, SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 2, SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 3 | | Authors: | Jeyaprakash, A.A, Santamaria, A, Jayachandran, U, Chan, Y.W, Benda, C, Nigg, E.A, Conti, E. | | Deposit date: | 2012-02-15 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural and Functional Organization of the Ska Complex, a Key Component of the Kinetochore-Microtubule Interface.
Mol.Cell, 46, 2012
|
|
4A0I
 
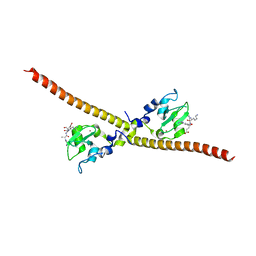 | | Crystal structure of Survivin bound to the N-terminal tail of hSgo1 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, SHUGOSHIN-LIKE 1, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4A0J
 
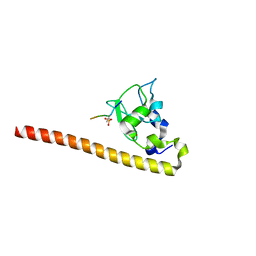 | | Crystal structure of Survivin bound to the phosphorylated N-terminal tail of histone H3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, HISTONE H3 PEPTIDE, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4A0N
 
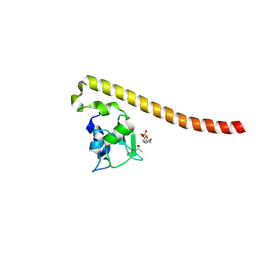 | | Crystal structure of Survivin bound to the phosphorylated N-terminal tail of histone H3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, HISTONE H3 PEPTIDE, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4C9Y
 
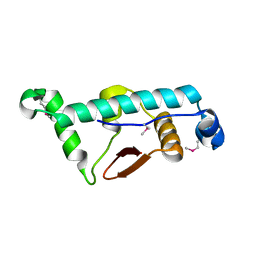 | | Structural Basis for the microtubule binding of the human kinetochore Ska complex | | Descriptor: | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1 | | Authors: | Abad, M, Medina, B, Santamaria, A, Zou, J, Plasberg-Hill, C, Madhumalar, A, Jayachandran, U, Redli, P.M, Rappsilber, J, Nigg, E.A, Jeyaprakash, A.A. | | Deposit date: | 2013-10-04 | | Release date: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for Microtubule Recognition by the Human Kinetochore Ska Complex.
Nat.Commun., 5, 2014
|
|
4CA0
 
 | | Structural Basis for the microtubule binding of the human kinetochore Ska complex | | Descriptor: | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1 | | Authors: | Abad, M, Medina, B, Santamaria, A, Zou, J, Plasberg-Hill, C, Madhumalar, A, Jayachandran, U, Redli, P.M, Rappsilber, J, Nigg, E.A, Jeyaprakash, A.A. | | Deposit date: | 2013-10-04 | | Release date: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.259 Å) | | Cite: | Structural Basis for Microtubule Recognition by the Human Kinetochore Ska Complex.
Nat.Commun., 5, 2014
|
|
2XB2
 
 | | Crystal structure of the core Mago-Y14-eIF4AIII-Barentsz-UPF3b assembly shows how the EJC is bridged to the NMD machinery | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Buchwald, G, Ebert, J, Basquin, C, Sauliere, J, Jayachandran, U, Bono, F, Le Hir, H, Conti, E. | | Deposit date: | 2010-04-03 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights Into the Recruitment of the Nmd Machinery from the Crystal Structure of a Core Ejc-Upf3B Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XZP
 
 | | Upf1 helicase | | Descriptor: | GLYCEROL, MALONATE ION, REGULATOR OF NONSENSE TRANSCRIPTS 1 | | Authors: | Chakrabarti, S, Jayachandran, U, Bonneau, F, Fiorini, F, Basquin, C, Domcke, S, Le Hir, H, Conti, E. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and its Regulation by Upf2.
Mol.Cell, 41, 2011
|
|
2XZL
 
 | | Upf1-RNA complex | | Descriptor: | 5- R(*UP*UP*UP*UP*UP*UP*UP*UP*U) -3, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HELICASE NAM7, ... | | Authors: | Chakrabarti, S, Jayachandran, U, Bonneau, F, Fiorini, F, Basquin, C, Domcke, S, Le Hir, H, Conti, E. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and its Regulation by Upf2.
Mol.Cell, 41, 2011
|
|
2XZO
 
 | | Upf1 helicase - RNA complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chakrabarti, S, Jayachandran, U, Bonneau, F, Fiorini, F, Basquin, C, Domcke, S, Le Hir, H, Conti, E. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and its Regulation by Upf2.
Mol.Cell, 41, 2011
|
|
