1DY6
 
 | | Structure of the imipenem-hydrolyzing beta-lactamase SME-1 | | Descriptor: | CARBAPENEM-HYDROLYSING BETA-LACTAMASE SME-1 | | Authors: | Sougakoff, W, L'Hermite, G, Billy, I, Guillet, V, Naas, T, Nordman, P, Jarlier, V, Delettre, J. | | Deposit date: | 2000-01-27 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of the Imipenem-Hydrolyzing Class a Beta-Lactamase Sme-1 from Serratia Marcescens.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1H8Y
 
 | | Crystal structure of the class D beta-lactamase OXA-13 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, BETA-LACTAMASE, SULFATE ION | | Authors: | Pernot, L, Frenois, F, Rybkine, T, L'Hermite, G, Petrella, S, Delettre, J, Jarlier, V, Collatz, E, Sougakoff, W. | | Deposit date: | 2001-02-17 | | Release date: | 2001-07-12 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Class D B-Lactamase Oxa-13 in the Native Form and in Complex with Meropenem
J.Mol.Biol., 310, 2001
|
|
1H8Z
 
 | | Crystal structure of the class D beta-lactamase OXA-13 | | Descriptor: | BETA-LACTAMASE, SULFATE ION | | Authors: | Pernot, L, Frenois, F, Rybkine, T, L'Hermite, G, Petrella, S, Delettre, J, Jarlier, V, Collatz, E, Sougakoff, W. | | Deposit date: | 2001-02-17 | | Release date: | 2001-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Class D B-Lactamase Oxa-13 in the Native Form and in Complex with Meropenem
J.Mol.Biol., 310, 2001
|
|
3C5A
 
 | | Crystal structure of the C-terminal deleted mutant of the class A carbapenemase KPC-2 at 1.23 angstrom | | Descriptor: | CITRIC ACID, Class A carbapenemase KPC-2 | | Authors: | Petrella, S, Ziental-Gelus, N, Mayer, C, Jarlier, V, Sougakoff, W. | | Deposit date: | 2008-01-31 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Genetic and structural insights into the dissemination potential of the extremely-broad-spectrum class A {beta}-lactamase (EBSBL) KPC-2 identified in two strains of Escherichia coli and Enterobacter cloacae isolated from the same patient in France
ANTIMICROB.AGENTS CHEMOTHER., 2008
|
|
3DW0
 
 | | Crystal structure of the class A carbapenemase KPC-2 at 1.6 angstrom resolution | | Descriptor: | Class A carbapenemase KPC-2 | | Authors: | Petrella, S, Ziental-Gelus, N, Mayer, C, Jarlier, V, Sougakoff, W. | | Deposit date: | 2008-07-21 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A beta-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France.
Antimicrob.Agents Chemother., 52, 2008
|
|
3M4I
 
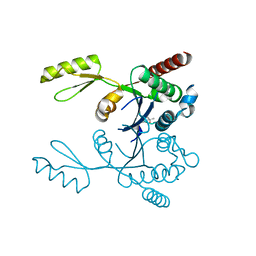 | | Crystal structure of the second part of the Mycobacterium tuberculosis DNA gyrase reaction core: the TOPRIM domain at 1.95 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit B | | Authors: | Piton, J, Petrella, S, Aubry, A, Mayer, C. | | Deposit date: | 2010-03-11 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase.
Plos One, 5, 2010
|
|
1O7E
 
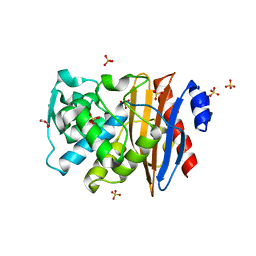 | |
3IG0
 
 | |
3IFZ
 
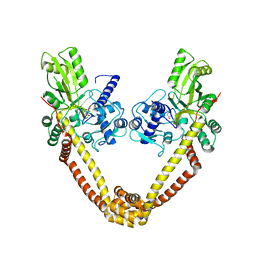 | | crystal structure of the first part of the Mycobacterium tuberculosis DNA gyrase reaction core: the breakage and reunion domain at 2.7 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit A, SODIUM ION | | Authors: | Piton, J, Aubry, A, Delarue, M, Mayer, C. | | Deposit date: | 2009-07-27 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase.
Plos One, 5, 2010
|
|
