6ELD
 
 | |
6SLO
 
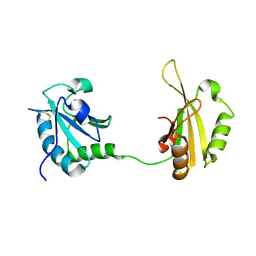 | | Crystal structure of PUF60 UHM domain in complex with 7,8 dimethoxyperphenazine | | Descriptor: | 2-[4-[3-(8-chloranyl-2,3-dimethoxy-phenothiazin-10-yl)propyl]piperazin-1-yl]ethanol, MAGNESIUM ION, Thioredoxin,Poly(U)-binding-splicing factor PUF60 | | Authors: | Jagtap, P.K.A, Kubelka, T, Bach, T, Sattler, M. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of phenothiazine derivatives as UHM-binding inhibitors of early spliceosome assembly.
Nat Commun, 11, 2020
|
|
8PJB
 
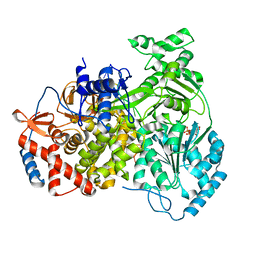 | |
8PJJ
 
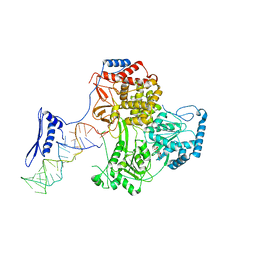 | |
4HCQ
 
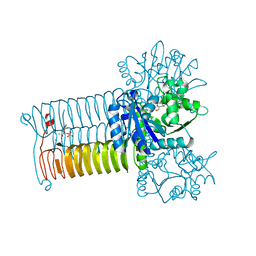 | | Crystal structure of GLMU from mycobacterium tuberculosis in complex with glucosamine-1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein GlmU, COBALT (II) ION, ... | | Authors: | Jagtap, P.K.A, Verma, S.K, Vithani, N. | | Deposit date: | 2012-10-01 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures identify an atypical two-metal-ion mechanism for uridyltransfer in GlmU: its significance to sugar nucleotidyl transferases
J.Mol.Biol., 425, 2013
|
|
8B9J
 
 | |
8B9L
 
 | |
8B9G
 
 | |
8B9K
 
 | |
8B9I
 
 | |
5O2V
 
 | | NMR structure of TIA-1 RRM1 domain | | Descriptor: | Nucleolysin TIA-1 isoform p40 | | Authors: | Jagtap, P.K.A. | | Deposit date: | 2017-05-22 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7QFT
 
 | | Crystal structure of KLK6 in complex with compound 16a | | Descriptor: | KLK6 Activity-Based Probe (Ahx-DPhe-Cha-Dht-Arg-DPP), Kallikrein-6 | | Authors: | Jagtap, P.K.A, Zhang, L, De Vita, E, Tate, E.W, Hennig, J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion.
J.Am.Chem.Soc., 144, 2022
|
|
7QFV
 
 | | Crystal structure of KLK6 in complex with compound 17a | | Descriptor: | KLK6 Activity-Based Probe (Ahx-DPhe-Ser(Z)-Dht-Arg-DPP), Kallikrein-6 | | Authors: | Jagtap, P.K.A, Zhang, L, De Vita, E, Tate, E.W, Hennig, J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion.
J.Am.Chem.Soc., 144, 2022
|
|
7QI0
 
 | | Crystal structure of KLK6 in complex with compound DKFZ918 | | Descriptor: | (5~{R})-3-(6-carbamimidoylpyridin-3-yl)-~{N}-[(1~{S})-1-naphthalen-1-ylpropyl]-2-oxidanylidene-1,3-oxazolidine-5-carboxamide, Kallikrein-6 | | Authors: | Jagtap, P.K.A, Baumann, A, Lohbeck, J, Isak, D, Miller, A, Hennig, J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Scalable synthesis and structural characterization of reversible KLK6 inhibitors.
Rsc Adv, 12, 2022
|
|
7QHZ
 
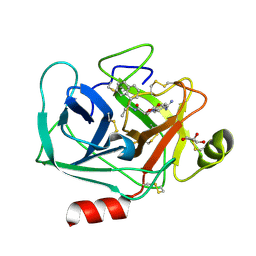 | | Crystal structure of KLK6 in complex with compound DKFZ917 | | Descriptor: | (5~{R})-3-(4-carbamimidoylphenyl)-~{N}-[(1~{S})-1-naphthalen-1-ylpropyl]-2-oxidanylidene-1,3-oxazolidine-5-carboxamide, GLYCEROL, Kallikrein-6 | | Authors: | Jagtap, P.K.A, Baumann, A, Lohbeck, J, Isak, D, Miller, A, Hennig, J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Scalable synthesis and structural characterization of reversible KLK6 inhibitors.
Rsc Adv, 12, 2022
|
|
7ZHH
 
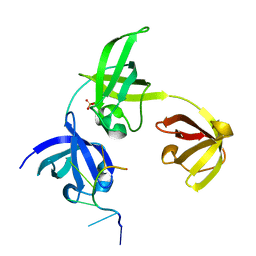 | | Complex structure of drosophila Unr CSD789 and a poly(A) RNA sequence | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, Upstream of N-ras, ... | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res., 51, 2023
|
|
7ZHR
 
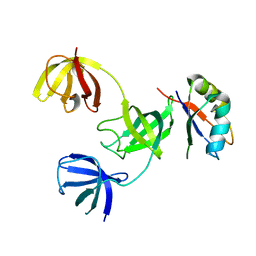 | |
5NV8
 
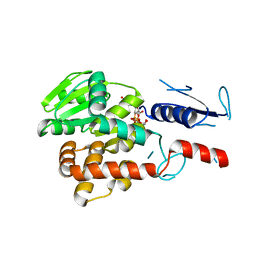 | | Structural basis for EarP-mediated arginine glycosylation of translation elongation factor EF-P | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, EF-P arginine 32 rhamnosyl-transferase | | Authors: | Macosek, J, Krafczyk, R, Jagtap, P.K.A, Lassaka, J, Hennig, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-10-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural Basis for EarP-Mediated Arginine Glycosylation of Translation Elongation Factor EF-P.
MBio, 8, 2017
|
|
5O3J
 
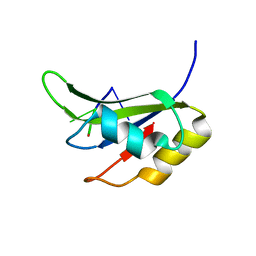 | | Crystal structure of TIA-1 RRM2 in complex with RNA | | Descriptor: | Nucleolysin TIA-1 isoform p40, RNA (5'-R(P*UP*UP*C)-3') | | Authors: | Sonntag, M, Jagtap, P.K.A, Hennig, J, Sattler, M. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7BJS
 
 | | Crystal structure of Khc/atypical Tm1 complex | | Descriptor: | Kinesin heavy chain, SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
7BJN
 
 | | Crystal structure of atypical Tm1 (Tm1-I/C), residues 270-334 | | Descriptor: | SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
7BJG
 
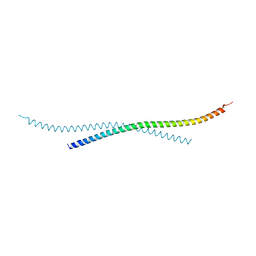 | | Crystal structure of atypical Tm1 (Tm1-I/C), residues 262-363 | | Descriptor: | SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
6Y6E
 
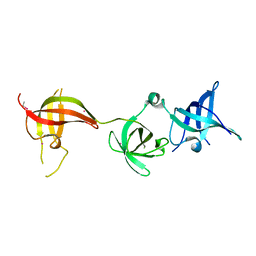 | | drosophila Unr CSD456 | | Descriptor: | ETHYL MERCURY ION, Upstream of N-ras, isoform A | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-29 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras.
Cell Rep, 32, 2020
|
|
7PCV
 
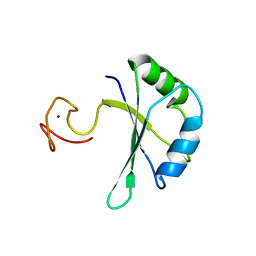 | |
7PDV
 
 | |
