3FEA
 
 | |
3FE7
 
 | |
6HIK
 
 | |
6Q9L
 
 | |
6Q9U
 
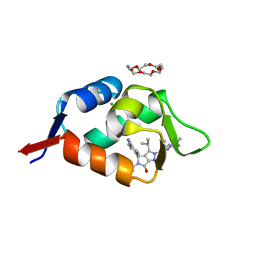 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 12 AT 2.4A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-[(4~{S})-5-(5-chloranyl-2-oxidanylidene-1~{H}-pyridin-3-yl)-2-[2-(dimethylamino)-4-methoxy-pyrimidin-5-yl]-6-oxidanylidene-3-propan-2-yl-4~{H}-pyrrolo[3,4-c]pyrazol-4-yl]benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9O
 
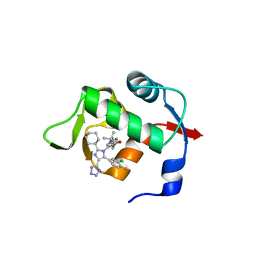 | |
6Q9Q
 
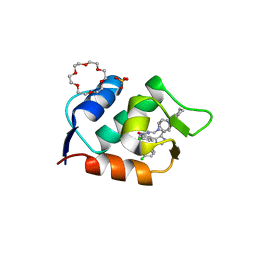 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 13 AT 2.1A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 6-chloranyl-3-[3-[(1~{S})-1-(4-chlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-~{N}-[2-(4-cyclohexylpiperazin-1-yl)ethyl]-1~{H}-indole-2-carboxamide, GLYCEROL, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9W
 
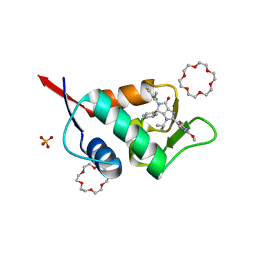 | | X-ray structure of compound 15 bound to HdmX: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | (4~{S})-4-(4-chlorophenyl)-5-[(1~{S})-1-(3-chlorophenyl)ethyl]-2-(2,4-dimethoxypyrimidin-5-yl)-3-propan-2-yl-4~{H}-pyrrolo[3,4-d]imidazol-6-one, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9H
 
 | |
6Q9Y
 
 | |
6Q9S
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 14 AT 2.4A: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[[6-chloranyl-3-[3-[(1~{S})-1-(2,4-dichlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-1~{H}-indol-2-yl]carbonylamino]-4-[4-(2-oxidanylidene-1,3-oxazinan-3-yl)piperidin-1-yl]benzoic acid, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q96
 
 | |
6YF1
 
 | | FKBP12 in complex with the BMP potentiator compound 8 at 1.12A resolution | | Descriptor: | (1aR,3R,5S,6R,7S,9R,10R,17aS,20S,21R,22S,25R,25aR)-25-Ethyl-10,22-dihydroxy-20-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-5,7-dimethoxy-1a,3,9,21-tetramethyloctadecahydro-2H-6,10-epoxyoxireno[p]pyrido[2,1-c][1,4]oxazacyclotricosine-11,12,18,24(1aH,14H)-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF3
 
 | | FKBP12 in complex with the BMP potentiator compound 10 at 1.00A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-17-ethyl-25-methoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14,23-tris(oxidanyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF2
 
 | | FKBP12 in complex with the BMP potentiator compound 6 at 1.03A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-23,25-dimethoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14-bis(oxidanyl)-17-(2-oxidanylidenepropyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF0
 
 | | FKBP12 in complex with the BMP potentiator compound 9 at 1.55 A resolution | | Descriptor: | 18-HYDROXYASCOMYCIN, Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
3KGA
 
 | | Crystal structure of MAPKAP kinase 2 (MK2) complexed with a potent 3-aminopyrazole ATP site inhibitor | | Descriptor: | 6-{3-amino-1-[3-(1H-indol-6-yl)phenyl]-1H-pyrazol-4-yl}-3,4-dihydroisoquinolin-1(2H)-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Kroemer, M, Velcicky, J, Izaac, A, Be, C, Huppertz, C, Pflieger, D, Schlapbach, A, Scheufler, C. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Novel 3-aminopyrazole inhibitors of MK-2 discovered by scaffold hopping strategy.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3M2W
 
 | | Crystal structure of MAPKAK kinase 2 (MK2) complexed with a spiroazetidine-tetracyclic ATP site inhibitor | | Descriptor: | 2'-(2-fluorophenyl)-1-methyl-6',8',9',11'-tetrahydrospiro[azetidine-3,10'-pyrido[3',4':4,5]pyrrolo[2,3-f]isoquinolin]-7'(5'H)-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Kroemer, M, Revesz, L, Be, C, Izaac, A, Huppertz, C, Schlapbach, A, Scheufler, C. | | Deposit date: | 2010-03-08 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | In vivo and in vitro SAR of tetracyclic MAPKAP-K2 (MK2) inhibitors. Part II.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3M42
 
 | | Crystal structure of MAPKAP kinase 2 (MK2) complexed with a tetracyclic ATP site inhibitor | | Descriptor: | 2-[5-(2-methoxyethoxy)pyridin-3-yl]-8,9,10,11-tetrahydro-7H-pyrido[3',4':4,5]pyrrolo[2,3-f]isoquinolin-7-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Scheufler, C, Revesz, L, Be, C, Izaac, A, Huppertz, C, Schlapbach, A, Kroemer, M. | | Deposit date: | 2010-03-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Novel pyrrolo[2,3-f]isoquinoline based MAPKAP-K2 (MK2) inhibitors with potent in vitro and in vivo activity
To be Published
|
|
5HHW
 
 | | Crystal structure of insulin receptor kinase domain in complex with cis-(R)-7-(3-(azetidin-1-ylmethyl)cyclobutyl)-5-(3-((tetrahydro-2H-pyran-2-yl)methoxy)phenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine. | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-(azetidin-1-ylmethyl)cyclobutyl]-5-[3-[[(2~{R})-oxan-2-yl]methoxy]phenyl]pyrrolo[2,3-d]pyrimidin-4-amine, Insulin receptor | | Authors: | Scheufler, C, Izaac, A, Stauffer, F. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-13 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of a 5-[3-phenyl-(2-cyclic-ether)-methylether]-4-aminopyrrolo[2,3-d]pyrimidine series of IGF-1R inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6FYK
 
 | | X-Ray structure of CLK2-KD(136-496)/Indazole1 at 2.39A | | Descriptor: | (2~{S})-4-methyl-1-[5-(3-methyl-2~{H}-indazol-5-yl)pyridin-3-yl]oxy-pentan-2-amine, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYL
 
 | | X-ray structure of CLK2-KD(136-496)/CX-4945 at 1.95A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYV
 
 | | X-RAY STRUCTURE OF CLK4-KD(146-480)/CX-4945 AT 2.46A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK4, SULFATE ION | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYO
 
 | | X-RAY STRUCTURE OF CLK1-KD(148-484)/Cpd-2 AT 2.32A | | Descriptor: | 6-~{tert}-butyl-~{N}-[6-(1~{H}-pyrazol-4-yl)-1~{H}-imidazo[1,2-a]pyridin-2-yl]pyridine-3-carboxamide, Dual specificity protein kinase CLK1, SULFATE ION | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYP
 
 | | X-RAY STRUCTURE OF CLK3-KD(GP-[275-632], NON-PHOS.)/CX-4945 AT 2.29A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK3 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
