2MX1
 
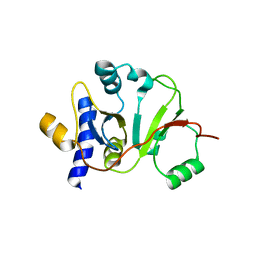 | | Structure of the E. coli Threonylcarbamoyl-AMP Synthase TSAC | | Descriptor: | Threonylcarbamoyl-AMP synthase | | Authors: | Harris, K.A, Bobay, B.G, Sarachan, K.L, Sims, A.F, Bilbille, Y, Deutsch, C, Iwata-Reuyl, D, Agris, P.F. | | Deposit date: | 2014-12-06 | | Release date: | 2015-06-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR-based Structural Analysis of Threonylcarbamoyl-AMP Synthase and Its Substrate Interactions.
J.Biol.Chem., 290, 2015
|
|
5UDG
 
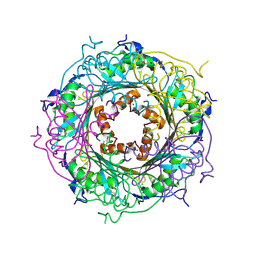 | | Mutant E97Q crystal structure of Bacillus subtilis QueF with a disulfide Cys 55-99 | | Descriptor: | MAGNESIUM ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, TRIETHYLENE GLYCOL | | Authors: | Mohammad, A, Kiani, M.K, Iwata-Reuyl, D, Stec, B, Swairjo, M. | | Deposit date: | 2016-12-27 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protection of the Queuosine Biosynthesis Enzyme QueF from Irreversible Oxidation by a Conserved Intramolecular Disulfide.
Biomolecules, 7, 2017
|
|
3D2O
 
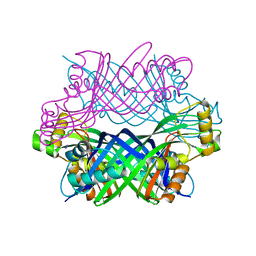 | | Crystal Structure of Manganese-metallated GTP Cyclohydrolase Type IB | | Descriptor: | AZIDE ION, CHLORIDE ION, LITHIUM ION, ... | | Authors: | Swairjo, M.A. | | Deposit date: | 2008-05-08 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB
J.Bacteriol., 191, 2009
|
|
5K9G
 
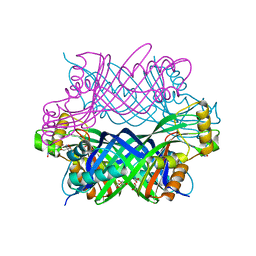 | | Crystal Structure of GTP Cyclohydrolase-IB with Tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Alvarez, J, Stec, B, Swairjo, M.A. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism and catalytic strategy of the prokaryotic-specific GTP cyclohydrolase-IB.
Biochem.J., 474, 2017
|
|
6N9A
 
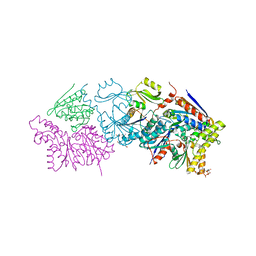 | |
8DL3
 
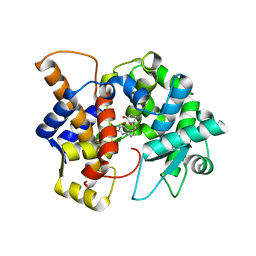 | | Crystal structure of the human queuine salvage enzyme DUF2419, complexed with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, Queuosine salvage protein | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-07-06 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
5CAJ
 
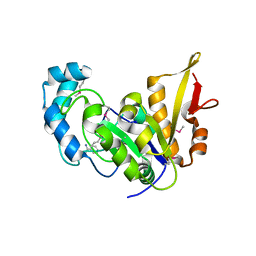 | | Crystal structure of E. coli YaaA, a member of the DUF328/UPF0246 family | | Descriptor: | BENZAMIDINE, CHLORIDE ION, UPF0246 protein YaaA | | Authors: | Prahlad, J, Lin, J, Wilson, M.A. | | Deposit date: | 2015-06-29 | | Release date: | 2016-02-24 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The DUF328 family member YaaA is a DNA-binding protein with a novel fold.
J.Biol.Chem., 295, 2020
|
|
5JYX
 
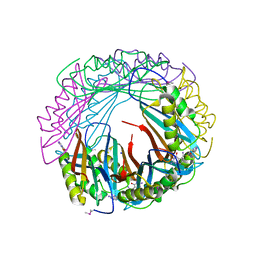 | |
5K0P
 
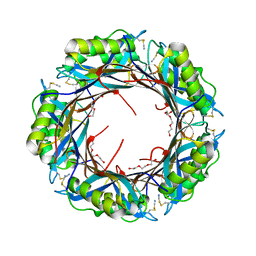 | |
5K95
 
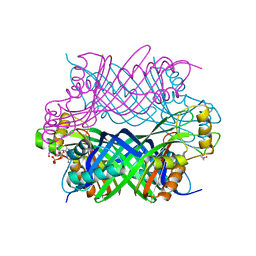 | |
4F8B
 
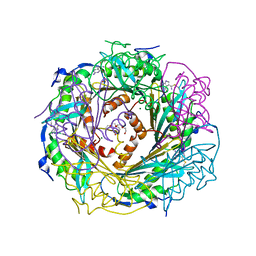 | | Crystal Structure of the Covalent Thioimide Intermediate of Unimodular Nitrile Reductase QueF | | Descriptor: | 2-amino-5-[(Z)-iminomethyl]-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, MAGNESIUM ION, ... | | Authors: | Stec, B, Swairjo, M.A. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis of biological nitrile reduction.
J.Biol.Chem., 287, 2012
|
|
4FGC
 
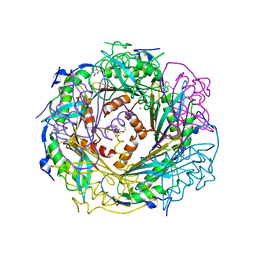 | | Crystal Structure of Active Site Mutant C55A of Nitrile Reductase QueF, Bound to Substrate PreQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, ... | | Authors: | Stec, B, Swairjo, M.A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structural basis of biological nitrile reduction.
J.Biol.Chem., 287, 2012
|
|
7UGK
 
 | |
7U1O
 
 | | Crystal structure of queuine salvage enzyme DUF2419 complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7U91
 
 | | Crystal structure of queuine salvage enzyme DUF2419, in complex with queuosine-5'-monophosphate | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-(5-O-phosphono-beta-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7U07
 
 | |
7U5A
 
 | | Crystal structure of queuine salvage enzyme DUF2419 mutant K199C, complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, MALONATE ION, Queuine salvage enzyme DUF2419 | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7UK3
 
 | |
7UI4
 
 | |
7ULC
 
 | | Crystal structure of queuine salvage enzyme DUF2419 mutant D231N, in complex with queuosine-5'-monophosphate | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-(5-O-phosphono-beta-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, Queuosine salvage protein DUF2419 | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-04-04 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
