1AA9
 
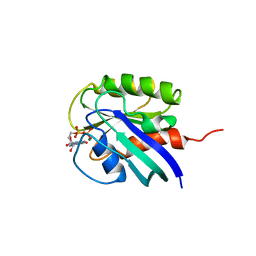 | | HUMAN C-HA-RAS(1-171)(DOT)GDP, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | C-HA-RAS, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Ito, Y, Yamasaki, Y, Muto, Y, Kawai, G, Nishimura, S, Miyazawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-01-27 | | Release date: | 1997-07-29 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein.
Biochemistry, 36, 1997
|
|
8JYH
 
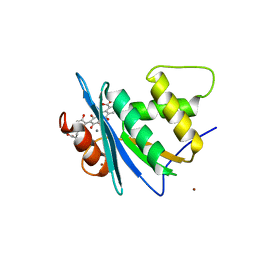 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid C | | Descriptor: | 7-[5-[(2~{S})-2-azanyl-3-oxidanyl-3-oxidanylidene-propyl]-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,HIV-1 Reverse Transcriptase RNase H active domain, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
8JYJ
 
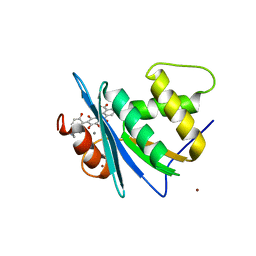 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid A | | Descriptor: | 7-[5-(2-acetamidoethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,HIV-1 Reverse Transcriptase RNase H active domain, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
8JYI
 
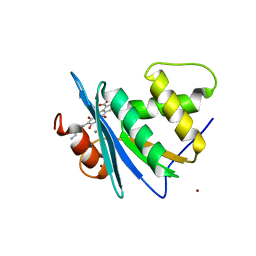 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid E | | Descriptor: | 7-[5-(2-azanylethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,Ribonuclease H, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
7FAO
 
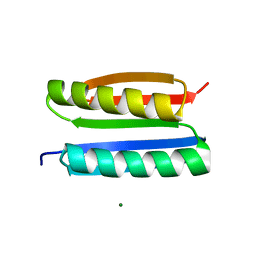 | |
1WVO
 
 | | Solution structure of RSGI RUH-029, an antifreeze protein like domain in human N-acetylneuraminic acid phosphate synthase gene. | | Descriptor: | Sialic acid synthase | | Authors: | Ito, Y, Hamada, T, Hayashi, F, Yokoyama, S, Hirota, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-22 | | Release date: | 2006-01-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antifreeze-like domain of human sialic acid synthase
Protein Sci., 15, 2006
|
|
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N9L
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in-cell GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
8AU4
 
 | | Structural insights reveal a heterotetramer between oncogenic K-Ras4BG12V and Rgl2, a RalA/B activator | | Descriptor: | Ral guanine nucleotide dissociation stimulator-like 2 | | Authors: | Tariq, M, Ikeya, T, Togashi, N, Fairall, L, Alejo, C.B, Kamei, S, Alonso, B.R, Campillo, M.A.M, Hudson, A, Ito, Y, Schwabe, J, Dominguez, C, Tanaka, K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
1X3U
 
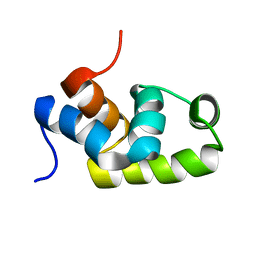 | | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium melilot | | Descriptor: | Transcriptional regulatory protein fixJ | | Authors: | Kurashima-Ito, K, Kasai, Y, Hosono, K, Tamura, K, Oue, S, Isogai, M, Ito, Y, Nakamura, H, Shiro, Y. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium meliloti and its recognition of the fixK promoter
Biochemistry, 44, 2005
|
|
2CW1
 
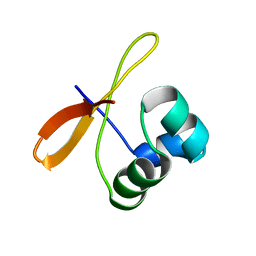 | | Solution structure of the de novo-designed lambda Cro fold protein | | Descriptor: | SN4m | | Authors: | Isogai, Y, Ito, Y, Ikeya, T, Shiro, Y, Ota, M. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Design of lambda Cro fold: solution structure of a monomeric variant of the de novo protein.
J.Mol.Biol., 354, 2005
|
|
1FJD
 
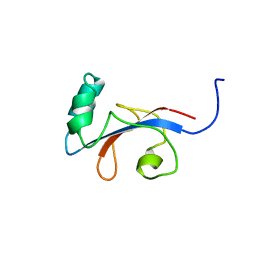 | | HUMAN PARVULIN-LIKE PEPTIDYL PROLYL CIS/TRANS ISOMERASE, HPAR14 | | Descriptor: | PEPTIDYL PROLYL CIS/TRANS ISOMERASE (PPIASE) | | Authors: | Terada, T, Shirouzu, M, Fukumori, Y, Fujimori, F, Ito, Y, Kigawa, T, Yokoyama, S, Uchida, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human parvulin-like peptidyl prolyl cis/trans isomerase, hPar14.
J.Mol.Biol., 305, 2001
|
|
2ROG
 
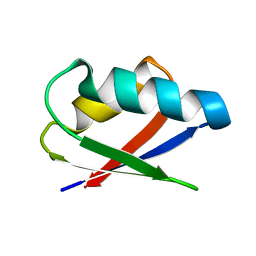 | | Solution structure of Thermus thermophilus HB8 TTHA1718 protein in living E. coli cells | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-03 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
2RRD
 
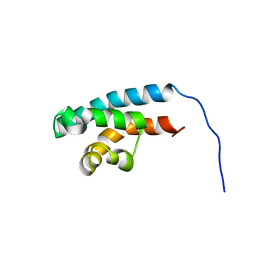 | | Structure of HRDC domain from human Bloom syndrome protein, BLM | | Descriptor: | HRDC domain from Bloom syndrome protein | | Authors: | Sato, A, Mishima, M, Nagai, A, Kim, S.Y, Ito, Y, Hakoshima, T, Jee, J.G, Kitano, K. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HRDC domain of human Bloom syndrome protein BLM
J.Biochem., 148, 2010
|
|
2RPH
 
 | | RecT-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
2RPD
 
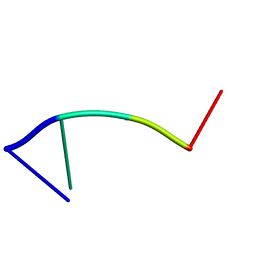 | | Mhr1p-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
2ROE
 
 | | Solution structure of thermus thermophilus HB8 TTHA1718 protein in vitro | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-20 | | Release date: | 2009-03-03 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
2RPF
 
 | | RecO-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
2RPE
 
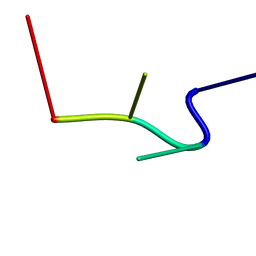 | | hsRad51-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
5WTQ
 
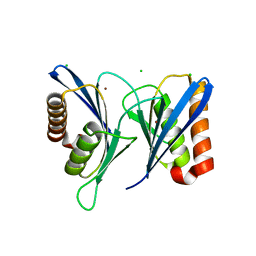 | | Crystal structure of human proteasome-assembling chaperone PAC4 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Proteasome assembly chaperone 4 | | Authors: | Kurimoto, E, Satoh, T, Ito, Y, Ishihara, E, Tanaka, K, Kato, K. | | Deposit date: | 2016-12-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human proteasome assembly chaperone PAC4 involved in proteasome formation
Protein Sci., 26, 2017
|
|
1AA3
 
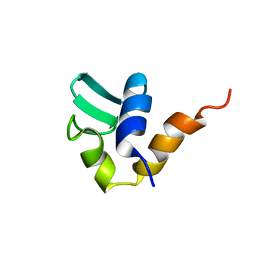 | | C-TERMINAL DOMAIN OF THE E. COLI RECA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RECA | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Terada, T, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing.
J.Mol.Biol., 274, 1997
|
|
1B22
 
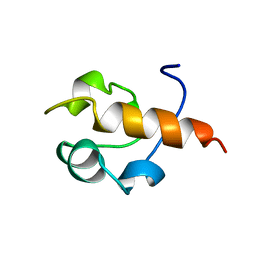 | | RAD51 (N-TERMINAL DOMAIN) | | Descriptor: | DNA REPAIR PROTEIN RAD51 | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR.
J.Mol.Biol., 290, 1999
|
|
1CMO
 
 | | IMMUNOGLOBULIN MOTIF DNA-RECOGNITION AND HETERODIMERIZATION FOR THE PEBP2/CBF RUNT-DOMAIN | | Descriptor: | POLYOMAVIRUS ENHANCER BINDING PROTEIN 2 | | Authors: | Nagata, T, Gupta, V, Sorce, D, Kim, W.Y, Sali, A, Chait, B.T, Shigesada, K, Ito, Y, Werner, M.H. | | Deposit date: | 1999-05-11 | | Release date: | 2000-01-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain.
Nat.Struct.Biol., 6, 1999
|
|
1CL3
 
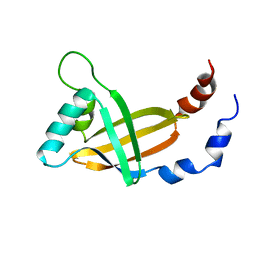 | | MOLECULAR INSIGHTS INTO PEBP2/CBF-SMMHC ASSOCIATED ACUTE LEUKEMIA REVEALED FROM THE THREE-DIMENSIONAL STRUCTURE OF PEBP2/CBF BETA | | Descriptor: | POLYOMAVIRUS ENHANCER BINDING PROTEIN 2 | | Authors: | Goger, M, Gupta, V, Kim, W.Y, Shigesada, K, Ito, Y, Werner, M.H. | | Deposit date: | 1999-05-04 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Molecular insights into PEBP2/CBF beta-SMMHC associated acute leukemia revealed from the structure of PEBP2/CBF beta
Nat.Struct.Biol., 6, 1999
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
