1D4U
 
 | | INTERACTIONS OF HUMAN NUCLEOTIDE EXCISION REPAIR PROTEIN XPA WITH RPA70 AND DNA: CHEMICAL SHIFT MAPPING AND 15N NMR RELAXATION STUDIES | | Descriptor: | NUCLEOTIDE EXCISION REPAIR PROTEIN XPA (XPA-MBD), ZINC ION | | Authors: | Buchko, G.W, Daughdrill, G.W, de Lorimier, R, Rao, S, Isern, N.G, Lingbeck, J, Taylor, J, Wold, M.S, Gochin, M, Spicer, L.D, Lowry, D.F, Kennedy, M.A. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies.
Biochemistry, 38, 1999
|
|
3QQN
 
 | | The retinal specific CD147 Ig0 domain: from molecular structure to biological activity | | Descriptor: | Basigin | | Authors: | Redzic, J.S, Armstrong, G.S, Isern, N.G, Kieft, J.S, Eisenmesser, E.Z, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2011-02-15 | | Release date: | 2011-05-11 | | Last modified: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | The Retinal Specific CD147 Ig0 Domain: From Molecular Structure to Biological Activity.
J.Mol.Biol., 411, 2011
|
|
3QR2
 
 | | Wild type CD147 Ig0 domain | | Descriptor: | Basigin | | Authors: | Redzic, J.S, Armstrong, G.S, Isern, N.G, Kieft, J.S, Eisenmesser, E.Z, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2011-02-16 | | Release date: | 2011-05-11 | | Last modified: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Retinal Specific CD147 Ig0 Domain: From Molecular Structure to Biological Activity.
J.Mol.Biol., 411, 2011
|
|
1SSV
 
 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-12-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
1SS7
 
 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-23 | | Release date: | 2004-12-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
2MVZ
 
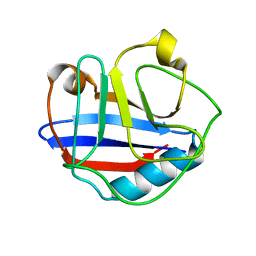 | | Solution Structure for Cyclophilin A from Geobacillus Kaustophilus | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Holliday, M.J, Isern, N.G, Geoffrey, A.S, Zhang, F, Eisenmesser, E.Z. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-08 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of GeoCyp: A Thermophilic Cyclophilin with a Novel Substrate Binding Mechanism That Functions Efficiently at Low Temperatures.
Biochemistry, 54, 2015
|
|
1EWI
 
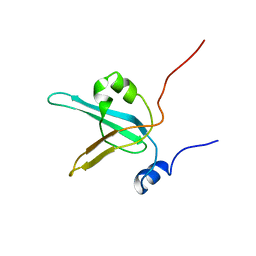 | | HUMAN REPLICATION PROTEIN A: GLOBAL FOLD OF THE N-TERMINAL RPA-70 DOMAIN REVEALS A BASIC CLEFT AND FLEXIBLE C-TERMINAL LINKER | | Descriptor: | REPLICATION PROTEIN A | | Authors: | Jacobs, D.M, Lipton, A.S, Isern, N.G, Daughdrill, G.W, Lowry, D.F, Gomes, X, Wold, M.S. | | Deposit date: | 2000-04-25 | | Release date: | 2000-05-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker.
J.Biomol.NMR, 14, 1999
|
|
2KKN
 
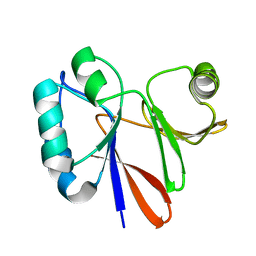 | | Solution NMR structure of Themotoga maritima protein TM1076: Northeast Structural Genomics Consortium target VT57 | | Descriptor: | Uncharacterized protein | | Authors: | Cort, J.R, Yee, A, Garcia, M, Isern, N.G, Arrowsmith, C.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-10-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Themotoga maritima protein TM1076
To be Published
|
|
2GJF
 
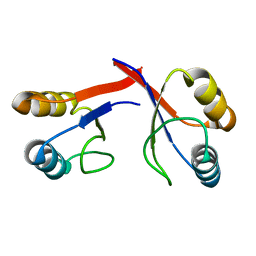 | |
2GJH
 
 | |
