1G80
 
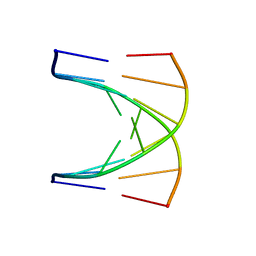 | | NMR SOLUTION STRUCTURE OF D(GCGTACGC)2 | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
1G7Z
 
 | | NMR SOLUTION STRUCTURE OF D(CGCTAGCG)2 | | Descriptor: | 5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
1KKW
 
 | |
1KKV
 
 | |
