1MTP
 
 | | The X-ray crystal structure of a serpin from a thermophilic prokaryote | | Descriptor: | Serine Proteinase Inhibitor (SERPIN), Chain A, Chain B | | Authors: | Irving, J.A, Cabrita, L.D, Rossjohn, J, Pike, R.N, Bottomley, S.P, Whisstock, J.C. | | Deposit date: | 2002-09-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A crystal structure of a prokaryote serpin: controlling conformational change in a heated environment
Structure, 11, 2003
|
|
1XU8
 
 | | The 2.8 A structure of a tumour suppressing serpin | | Descriptor: | Maspin, SULFATE ION | | Authors: | Irving, J.A, Law, R.H, Ruzyla, K, Bashtannyk-Puhalovich, T.A, Kim, N, Worrall, D.M, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
2H4R
 
 | |
8PI2
 
 | |
8P4U
 
 | |
8P4J
 
 | |
6GXX
 
 | | Fab fragment of an antibody selective for alpha-1-antitrypsin in the native conformation | | Descriptor: | FAB 1D9 heavy chain, FAB 1D9 light chain, MAGNESIUM ION | | Authors: | Elliston, E.L.K, Miranda, E, Perez, J, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-06-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterisation of a monoclonal antibody conformationally-selective for native alpha-1-antitrypsin
To Be Published
|
|
2DUT
 
 | |
6HX4
 
 | | Fab fragment of a native monomer-selective antibody in complex with alpha-1-antitrypsin | | Descriptor: | Alpha-1-antitrypsin, Fab 1D9 heavy chain, Fab 1D9 light chain | | Authors: | Elliston, E.L.K, Miranda, E, Perez, J, Jagger, A.M, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Characterisation of a monoclonal antibody conformationally-selective for native alpha-1-antitrypsin
To Be Published
|
|
6I1O
 
 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FAB 2H2 heavy chain, FAB 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
6I3Z
 
 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin in complex with its antigen | | Descriptor: | Alpha-1-antitrypsin, Fab 2H2 heavy chain, Fab 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Jagger, A.M, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
1SNG
 
 | | Structure of a Thermophilic Serpin in the Native State | | Descriptor: | COG4826: Serine protease inhibitor, SULFATE ION | | Authors: | Fulton, K.F, Buckle, A.M, Cabrita, L.D, Irving, J.A, Butcher, R.E, Smith, I, Reeve, S, Lesk, A.M, Bottomley, S.P, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2004-03-10 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition.
J.Biol.Chem., 280, 2005
|
|
4J1Y
 
 | | The X-ray crystal structure of human complement protease C1s zymogen | | Descriptor: | Complement C1s subcomponent | | Authors: | Perry, A.J, Wijeyewickrema, L.C, Wilmann, P.G, Gunzburg, M.J, D'Andrea, L, Irving, J.A, Pang, S.S, Duncan, R.C, Wilce, J.A, Whisstock, J.C, Pike, R.N. | | Deposit date: | 2013-02-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6645 Å) | | Cite: | A Molecular Switch Governs the Interaction between the Human Complement Protease C1s and Its Substrate, Complement C4.
J.Biol.Chem., 288, 2013
|
|
6I7U
 
 | |
6I4V
 
 | |
6IAY
 
 | |
1WZ9
 
 | | The 2.1 A structure of a tumour suppressing serpin | | Descriptor: | Maspin precursor, SULFATE ION | | Authors: | Law, R.H, Irving, J.A, Buckle, A.M, Ruzyla, K, Buzza, M, Bashtannyk-Puhalovich, T.A, Beddoe, T.C, Kim, N, Worrall, D.M, Bottomley, S.P, Bird, P.I, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2005-03-03 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
1YXA
 
 | | Serpina3n, a murine orthologue of human antichymotrypsin | | Descriptor: | serine (or cysteine) proteinase inhibitor, clade A, member 3N | | Authors: | Horvath, A.J, Irving, J.A, Law, R.H, Rossjohn, J, Bottomley, S.P, Quinsey, N.S, Pike, R.N, Coughlin, P.B, Whisstock, J.C. | | Deposit date: | 2005-02-20 | | Release date: | 2005-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins.
J.Biol.Chem., 280, 2005
|
|
6QU9
 
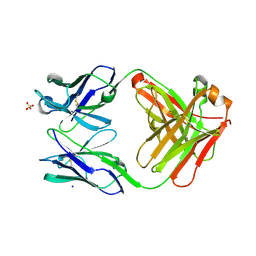 | | Fab fragment of an antibody that inhibits polymerisation of alpha-1-antitrypsin | | Descriptor: | FAB 4B12 heavy chain, FAB 4B12 light chain, GLYCEROL, ... | | Authors: | Jagger, A.M, Heyer-Chauhan, N, Lomas, D.A, Irving, J.A. | | Deposit date: | 2019-02-26 | | Release date: | 2020-03-18 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for Z alpha 1 -antitrypsin polymerization in the liver.
Sci Adv, 6, 2020
|
|
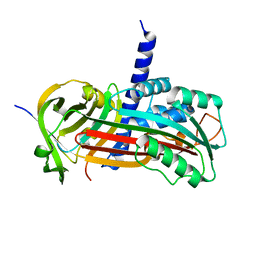
2H4Q
 
 | |
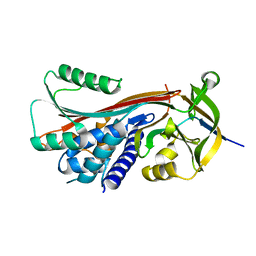
2H4P
 
 | |
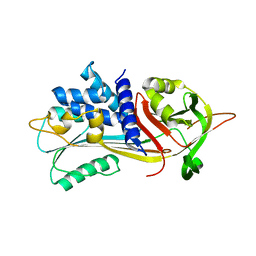
6ROD
 
 | |
7AEL
 
 | | alpha 1-antitrypsin (C232S) complexed with GSK716 | | Descriptor: | Alpha-1-antitrypsin, SULFATE ION, ~{N}-[(1~{S},2~{R})-1-(3-fluoranyl-2-methyl-phenyl)-1-oxidanyl-pentan-2-yl]-2-oxidanylidene-1,3-dihydroindole-4-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Development of a small molecule that corrects misfolding and increases secretion of Z alpha 1 -antitrypsin.
Embo Mol Med, 13, 2021
|
|
4PYW
 
 | | 1.92 angstrom crystal structure of A1AT:TTAI ternary complex | | Descriptor: | ACE-THR-THR-ALA-ILE-NH2, Alpha-1-antitrypsin, GLYCEROL | | Authors: | Nyon, M.P, Day, J, Gooptu, B. | | Deposit date: | 2014-03-28 | | Release date: | 2015-06-10 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An integrative approach combining ion mobility mass spectrometry, X-ray crystallography, and nuclear magnetic resonance spectroscopy to study the conformational dynamics of alpha 1 -antitrypsin upon ligand binding.
Protein Sci., 24, 2015
|
|
3G01
 
 | |
