2N5A
 
 | |
2N5B
 
 | |
2V4I
 
 | |
2VZK
 
 | | Structure of the acyl-enzyme complex of an N-terminal nucleophile (Ntn) hydrolase, OAT2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLUTAMATE N-ACETYLTRANSFERASE 2 ALPHA CHAIN, ... | | Authors: | Iqbal, A, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2008-08-01 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Anatomy of a Simple Acyl Intermediate in Enzyme Catalysis: Combined Biophysical and Modeling Studies on Ornithine Acetyl Transferase.
J.Am.Chem.Soc., 131, 2009
|
|
2WPW
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (without AcCoA) | | Descriptor: | ACETYL COENZYME *A, ORF14 | | Authors: | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | Deposit date: | 2009-08-11 | | Release date: | 2009-12-29 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
2WPX
 
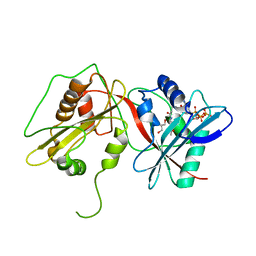 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (with AcCoA) | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, ORF14 | | Authors: | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | Deposit date: | 2009-08-11 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
5KE3
 
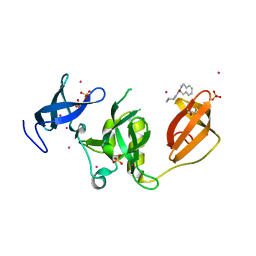 | | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a | | Descriptor: | (S)-N-(furan-2-ylmethyl)-1-(1,2,3,4-tetrahydroisoquinoline-3-carbonyl)piperidine-4-carboxamide, BETA-MERCAPTOETHANOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Ferreira de Freitas, R, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a
to be published
|
|
5KE2
 
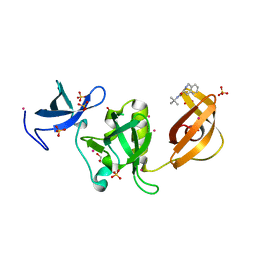 | | Crystal structure of SETDB1 Tudor domain in complex with inhibitor XST06472A | | Descriptor: | (3~{S})-~{N}-~{tert}-butyl-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with inhibitor xst06472a
to be published
|
|
2XFS
 
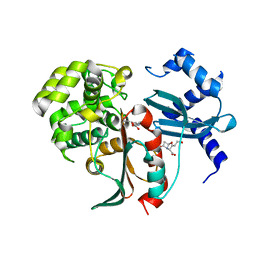 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XEP
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | CHLORIDE ION, GLYCEROL, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-17 | | Release date: | 2011-06-29 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XH9
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XGN
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | GLYCEROL, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-06-07 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XF3
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XFT
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | GLYCEROL, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4Z02
 
 | | Crystal structure of BRD1 in complex with Isoquinoline-3-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, UNKNOWN ATOM OR ION, ... | | Authors: | DONG, A, IQBAL, A, WALKER, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-25 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of BRD1 incomplex with Isoquinoline-3-carboxylic acid
to be published
|
|
2YEP
 
 | | STRUCTURE OF AN N-TERMINAL NUCLEOPHILE (NTN) HYDROLASE, OAT2, IN COMPLEX WITH GLUTAMATE | | Descriptor: | ACETATE ION, GLUTAMATE N-ACETYLTRANSFERASE 2 ALPHA CHAIN, GLUTAMATE N-ACETYLTRANSFERASE 2 BETA CHAIN, ... | | Authors: | Chowdhury, R, Iqbal, A, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2011-03-29 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Analyses Reveal How Ornithine Acetyl Transferase Binds Acidic and Basic Amino Acid Substrates.
Org.Biomol.Chem., 9, 2011
|
|
6AU2
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4H,6H-[1,2,4]triazolo[4,3-a][4,1]benzoxazepine, BETA-MERCAPTOETHANOL, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-10-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification and characterization of the first fragment hits for SETDB1 Tudor domain.
Bioorg.Med.Chem., 27, 2019
|
|
6AU3
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, N-{[2-(3,5-dimethyl-4H-1,2,4-triazol-4-yl)phenyl]methyl}acetamide, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments
to be published
|
|
4QQ6
 
 | | Crystal Structure of tudor domain of SMN1 in complex with a small organic molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
4QQD
 
 | | Crystal Structure of tandem tudor domains of UHRF1 in complex with a small organic molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, E3 ubiquitin-protein ligase UHRF1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
6BPI
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, MLY-SER-THR-E2G, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, DONG, A, DOBROVETSKY, E, IQBAL, A, CORLESS, V, TEMPEL, W, LIEW, S.K, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D.B, SCHAPIRA, M, VEDADI, M, BROWN, P.J, Santhakumar, V, FRYE, S, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates
to be published
|
|
6D6X
 
 | | HSP40 co-chaperone Sis1 J-domain | | Descriptor: | Type II HSP40 co-chaperone | | Authors: | Pinheiro, G.M.S, Amorim, G.C, Iqbal, A, Ramos, C.H.I, Almeida, F.C.L. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR investigation on the structure and function of the isolated J-domain from Sis1: Evidence of transient inter-domain interactions in the full-length protein.
Arch.Biochem.Biophys., 669, 2019
|
|
5BVN
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-[5-({[(3-fluorophenyl)carbamoyl]amino}methyl)-2-methylphenyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5BVK
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | 1-(2-chlorophenyl)-3-(pyridin-3-ylmethyl)urea, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5BVO
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(5-{(1S)-1-[(5-fluoro-1,3-benzoxazol-2-yl)amino]ethyl}-2-methylphenyl)imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
