1ED7
 
 | | SOLUTION STRUCTURE OF THE CHITIN-BINDING DOMAIN OF BACILLUS CIRCULANS WL-12 CHITINASE A1 | | Descriptor: | CHITINASE A1 | | Authors: | Ikegami, T, Okada, T, Hashimoto, M, Seino, S, Watanabe, T, Shirakawa, M. | | Deposit date: | 2000-01-27 | | Release date: | 2000-05-24 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1.
J.Biol.Chem., 275, 2000
|
|
1FR0
 
 | | SOLUTION STRUCTURE OF THE HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ANAEROBIC SENSOR KINASE ARCB FROM ESCHERICHIA COLI. | | Descriptor: | ARCB | | Authors: | Ikegami, T, Okada, T, Ohki, I, Hirayama, J, Mizuno, T, Shirakawa, M. | | Deposit date: | 2000-09-07 | | Release date: | 2001-03-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamic character of the histidine-containing phosphotransfer domain of anaerobic sensor kinase ArcB from Escherichia coli.
Biochemistry, 40, 2001
|
|
1XPA
 
 | | SOLUTION STRUCTURE OF THE DNA-AND RPA-BINDING DOMAIN OF THE HUMAN REPAIR FACTOR XPA, NMR, 1 STRUCTURE | | Descriptor: | XPA, ZINC ION | | Authors: | Ikegami, T, Kuraoka, I, Saijo, M, Kodo, N, Kyogoku, Y, Morikawa, K, Tanaka, K, Shirakawa, M. | | Deposit date: | 1998-07-06 | | Release date: | 1999-07-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA.
Nat.Struct.Biol., 5, 1998
|
|
1WSO
 
 | | The solution structures of human Orexin-A | | Descriptor: | Orexin-A | | Authors: | Ikegami, T, Takai, T. | | Deposit date: | 2004-11-08 | | Release date: | 2004-11-30 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Orexin-A is composed of a highly conserved C-terminal and a specific, hydrophilic N-terminal region, revealing the structural basis of specific recognition by the orexin-1 receptor
J.Pept.Sci., 12, 2006
|
|
5ZF0
 
 | | X-ray Structure of the Electron Transfer Complex between Ferredoxin and Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kubota-Kawai, H, Mutoh, R, Shinmura, K, Setif, P, Nowaczyk, M, Roegner, M, Ikegami, T, Tanaka, T, Kurisu, G. | | Deposit date: | 2018-03-01 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | X-ray structure of an asymmetrical trimeric ferredoxin-photosystem I complex
Nat Plants, 4, 2018
|
|
1K85
 
 | | Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 Chitinase A1. | | Descriptor: | CHITINASE A1 | | Authors: | Jee, J.G, Ikegami, T, Hashimoto, M, Kawabata, T, Ikeguchi, M, Watanabe, T, Shirakawa, M. | | Deposit date: | 2001-10-23 | | Release date: | 2002-12-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Fibronectin Type III Domain
from Bacillus circulans WL-12 Chitinase A1
J.Biol.Chem., 277, 2002
|
|
7WEM
 
 | | Solid-state NMR Structure of TFo c-Subunit Ring | | Descriptor: | ATP synthase subunit c | | Authors: | Akutsu, H, Todokoro, Y, Kang, S.-J, Suzuki, T, Yoshida, M, Ikegami, T, Fujiwara, T. | | Deposit date: | 2021-12-23 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-24 | | Method: | SOLID-STATE NMR | | Cite: | Chemical Conformation of the Essential Glutamate Site of the c -Ring within Thermophilic Bacillus F o F 1 -ATP Synthase Determined by Solid-State NMR Based on its Isolated c -Ring Structure.
J.Am.Chem.Soc., 144, 2022
|
|
1GEA
 
 | | RECEPTOR-BOUND CONFORMATION OF PACAP21 | | Descriptor: | PITUITARY ADENYLATE CYCLASE ACTIVATING POLYPEPTIDE | | Authors: | Inooka, H, Ohtaki, T, Kitahara, O, Ikegami, T, Endo, S, Kitada, C, Ogi, K, Onda, H, Fujino, M, Shirakawa, M. | | Deposit date: | 2000-10-20 | | Release date: | 2001-04-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Conformation of a peptide ligand bound to its G-protein coupled receptor.
Nat.Struct.Biol., 8, 2001
|
|
2ND5
 
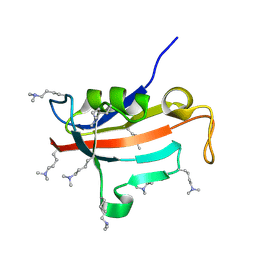 | | Lysine dimethylated FKBP12 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Hattori, Y, Sebera, J, Sychrovsky, V, Furuita, K, Sugiki, T, Ohki, I, Ikegami, T, Kobayashi, N, Tanaka, Y, Fujiwara, T, Kojima, C. | | Deposit date: | 2016-05-05 | | Release date: | 2017-05-17 | | Method: | SOLUTION NMR | | Cite: | NMR Observation of Protein Surface Salt Bridges at Neutral pH
To be Published
|
|
1SNH
 
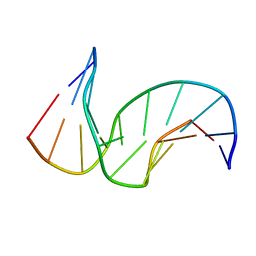 | | Solution structure of the DNA Decamer Duplex Containing Double TG Mismatches of Cis-syn Cyclobutane Pyrimidine Dimer | | Descriptor: | 5'-D(*CP*GP*CP*AP*TP*TP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*TP*GP*GP*TP*GP*CP*G)-3' | | Authors: | Lee, J.H, Park, C.J, Shin, J.S, Ikegami, T, Akutsu, H, Choi, B.S. | | Deposit date: | 2004-03-11 | | Release date: | 2004-05-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the DNA decamer duplex containing double T*G mismatches of cis-syn cyclobutane pyrimidine dimer: implications for DNA damage recognition by the XPC-hHR23B complex.
Nucleic Acids Res., 32, 2004
|
|
3A57
 
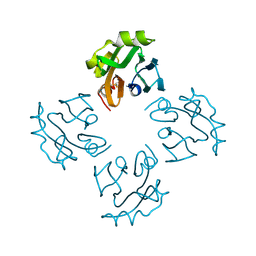 | | Crystal structure of Thermostable Direct Hemolysin | | Descriptor: | Thermostable direct hemolysin 2 | | Authors: | Hashimoto, H, Yanagihara, I, Nakahira, K, Hamada, D, Ikegami, T, Mayanagi, K, Kaieda, S, Fukui, T, Ohnishi, K, Kajiyama, S, Yamane, T, Ikeguchi, M, Honda, T, Shimizu, T, Sato, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin
J.Biol.Chem., 285, 2010
|
|
1IG4
 
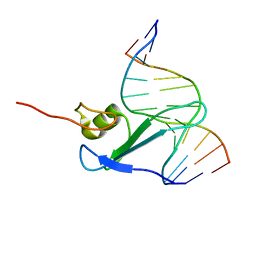 | | Solution Structure of the Methyl-CpG-Binding Domain of Human MBD1 in Complex with Methylated DNA | | Descriptor: | 5'-D(*GP*TP*AP*TP*CP*(5CM)P*GP*GP*AP*TP*AP*C)-3', Methyl-CpG Binding Protein | | Authors: | Ohki, I, Shimotake, N, Fujita, N, Jee, J.-G, Ikegami, T, Nakao, M, Shirakawa, M. | | Deposit date: | 2001-04-17 | | Release date: | 2001-05-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA.
Cell(Cambridge,Mass.), 105, 2001
|
|
1WU0
 
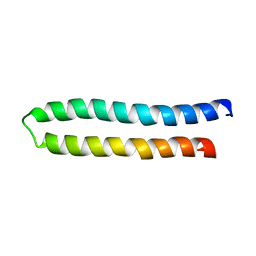 | |
1WUZ
 
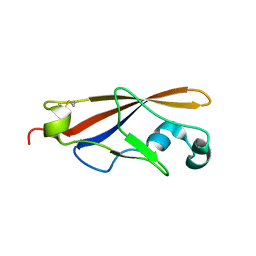 | | Structure of EC1 domain of CNR | | Descriptor: | Pcdha4 protein | | Authors: | Morishita, H, Umitsu, M, Yamaguchi, T, Murata, Y, Shibata, N, Udaka, K, Higuchi, Y, Akutsu, H, Yagi, T, Ikegami, T. | | Deposit date: | 2004-12-09 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural diversity of the first cadherin domains revealed by the structure of CNR/Protocadherin alpha
To be Published
|
|
2MXA
 
 | | Solution structure of the NDH-1 complex subunit CupS from Thermosynechococcus elongatus | | Descriptor: | NDH-1 complex sensory subunit CupS | | Authors: | Korste, A, Wulfhorst, H, Ikegami, T, Nowaczyk, M.M, Stoll, R. | | Deposit date: | 2014-12-17 | | Release date: | 2015-06-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the NDH-1 complex subunit CupS from Thermosynechococcus elongatus.
Biochim.Biophys.Acta, 1847, 2015
|
|
2D49
 
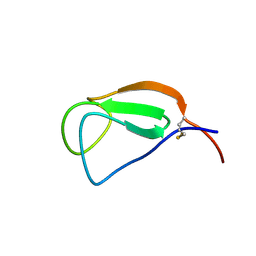 | | Solution structure of the Chitin-Binding Domain of Streptomyces griseus Chitinase C | | Descriptor: | chitinase C | | Authors: | Akagi, K, Watanabe, J, Hara, M, Kezuka, Y, Chikaishi, E, Yamaguchi, T, Akutsu, H, Nonaka, T, Watanabe, T, Ikegami, T. | | Deposit date: | 2005-10-11 | | Release date: | 2006-10-11 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Identification of the substrate interaction region of the chitin-binding domain of Streptomyces griseus chitinase C
J.Biochem.(Tokyo), 139, 2006
|
|
2CZN
 
 | | Solution structure of the chitin-binding domain of hyperthermophilic chitinase from pyrococcus furiosus | | Descriptor: | chitinase | | Authors: | Uegaki, T, Ikegami, T, Nakamura, T, Hagihara, Y, Mine, S, Inoue, T, Matsumura, H, Ataka, M, Ishikawa, K. | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-18 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus.
J.Mol.Biol., 381, 2008
|
|
2EIX
 
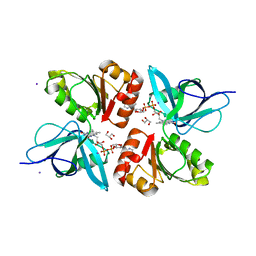 | | The Structure of Physarum polycephalum cytochrome b5 reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, IODIDE ION, ... | | Authors: | Kim, S.W, Suga, M, Ogasahara, K, Ikegami, T, Minami, Y, Yubisui, T, Tsukihara, T. | | Deposit date: | 2007-03-14 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of Physarum polycephalum cytochrome b5 reductase at 1.56 A resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | Descriptor: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | Authors: | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
2D1X
 
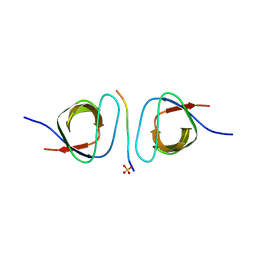 | | The crystal structure of the cortactin-SH3 domain and AMAP1-peptide complex | | Descriptor: | SULFATE ION, cortactin isoform a, proline rich region from development and differentiation enhancing factor 1 | | Authors: | Hashimoto, S, Hirose, M, Hashimoto, A, Morishige, M, Yamada, A, Hosaka, H, Akagi, K, Ogawa, E, Oneyama, C, Agatsuma, T, Okada, M, Kobayashi, H, Wada, H, Nakano, H, Ikegami, T, Nakagawa, A, Sabe, H. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting AMAP1 and cortactin binding bearing an atypical src homology 3/proline interface for prevention of breast cancer invasion and metastasis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2RUJ
 
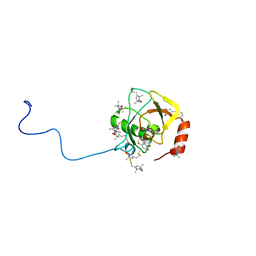 | | Solution structure of MTSL spin-labeled Schizosaccharomyces pombe Sin1 CRIM domain | | Descriptor: | Stress-activated map kinase-interacting protein 1 | | Authors: | Furuita, K, Kataoka, S, Sugiki, T, Kobayashi, N, Ikegami, T, Shiozaki, K, Fujiwara, T, Kojima, C. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Method: | SOLUTION NMR | | Cite: | Utilization of paramagnetic relaxation enhancements for high-resolution NMR structure determination of a soluble loop-rich protein with sparse NOE distance restraints
J.Biomol.Nmr, 61, 2015
|
|
2RTT
 
 | | Solution structure of the chitin-binding domain of Chi18aC from Streptomyces coelicolor | | Descriptor: | ChiC | | Authors: | Okumura, A, Uemura, M, Yamada, N, Chikaishi, E, Takai, T, Yoshio, S, Akagi, K, Morita, J, Lee, Y, Yokogawa, D, Suzuki, K, Watanabe, T, Ikegami, T. | | Deposit date: | 2013-08-26 | | Release date: | 2014-08-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Chitin-binding domain of chitinase Chi18aC from Streptomyces coelicolor
To be Published
|
|
2RRI
 
 | | NMR structure of vasoactive intestinal peptide in DPC Micelle | | Descriptor: | Vasoactive intestinal peptide | | Authors: | Umetsu, Y, Tenno, T, Goda, N, Shirakawa, M, Ikegami, T, Hiroaki, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural difference of vasoactive intestinal peptide in two distinct membrane-mimicking environments
Biochim.Biophys.Acta, 1814, 2011
|
|
2RSY
 
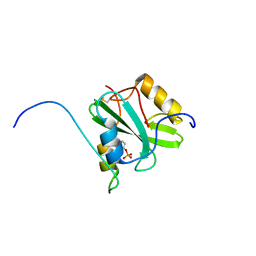 | | Solution structure of the SH2 domain of Csk in complex with a phosphopeptide from Cbp | | Descriptor: | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1, Tyrosine-protein kinase CSK | | Authors: | Tanaka, H, Akagi, K, Oneyama, C, Tanaka, M, Sasaki, Y, Kanou, T, Lee, Y, Yokogawa, D, Debenecker, M, Nakagawa, A, Okada, M, Ikegami, T. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-10 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Identification of a new interaction mode between the Src homology 2 domain of C-terminal Src kinase (Csk) and Csk-binding protein/phosphoprotein associated with glycosphingolipid microdomains.
J.Biol.Chem., 288, 2013
|
|
2RPQ
 
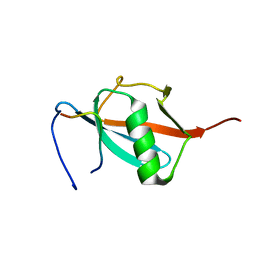 | | Solution Structure of a SUMO-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3 | | Descriptor: | Activating transcription factor 7-interacting protein 1, Small ubiquitin-related modifier 2 | | Authors: | Sekiyama, N, Ikegami, T, Yamane, T, Ikeguchi, M, Uchimura, Y, Baba, D, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2008-07-07 | | Release date: | 2008-10-07 | | Last modified: | 2015-12-09 | | Method: | SOLUTION NMR | | Cite: | Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3
J.Biol.Chem., 283, 2008
|
|
