1D0G
 
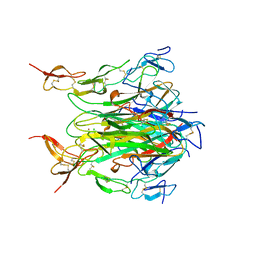 | | CRYSTAL STRUCTURE OF DEATH RECEPTOR 5 (DR5) BOUND TO APO2L/TRAIL | | Descriptor: | APOPTOSIS-2 LIGAND, CHLORIDE ION, DEATH RECEPTOR-5, ... | | Authors: | Hymowitz, S.G, Christinger, H.W, Fuh, G, O'Connell, M.P, Kelley, R.F, Ashkenazi, A, de Vos, A.M. | | Deposit date: | 1999-09-09 | | Release date: | 1999-10-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5.
Mol.Cell, 4, 1999
|
|
1DG6
 
 | | CRYSTAL STRUCTURE OF APO2L/TRAIL | | Descriptor: | APO2L/TNF-RELATED APOPOTIS INDUCING LIGAND (TRAIL), CHLORIDE ION, ZINC ION | | Authors: | Hymowitz, S.G, O'ConnelL, M.P, Ultsch, M.H, de Vos, A.M, Kelley, R.F. | | Deposit date: | 1999-11-23 | | Release date: | 2000-01-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A unique zinc-binding site revealed by a high-resolution X-ray structure of homotrimeric Apo2L/TRAIL.
Biochemistry, 39, 2000
|
|
1JPY
 
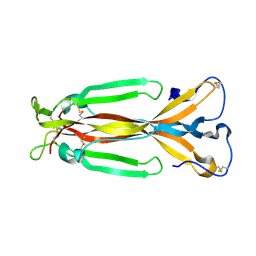 | | Crystal structure of IL-17F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hymowitz, S.G, Filvaroff, E.H, Yin, J, Lee, J, Cai, L, Risser, P, Maruoka, M, Mao, W, Foster, J, Kelley, R, Pan, G, Gurney, A.L, de Vos, A.M, Starovasnik, M.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding.
EMBO J., 20, 2001
|
|
1RJ8
 
 | | The crystal structure of TNF family member EDA-A2 | | Descriptor: | ectodysplasin-A isoform EDA-A2 | | Authors: | Hymowitz, S.G, Compaan, D.M, Yan, M, Ackerly, H, Dixit, V.M, Starovasnik, M.A, de Vos, A.M. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Crystal Structure of EDA-A1 and EDA-A2: splice variants with distinct receptor specificity
STRUCTURE, 11, 2003
|
|
1RJ7
 
 | | Crystal structure of EDA-A1 | | Descriptor: | Ectodysplasin A | | Authors: | Hymowitz, S.G, Compaan, D.M, Yan, M, Ackerly, H, Dixit, V.M, Starovasnik, M.A, de Vos, A.M. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structures of EDA-A1 and EDA-A2: splice variants with distinct receptor specificity.
Structure, 11, 2003
|
|
1XUT
 
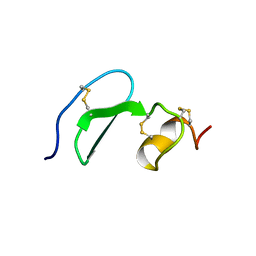 | | Solution structure of TACI-CRD2 | | Descriptor: | Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding.
J.Biol.Chem., 280, 2005
|
|
1XU1
 
 | | The crystal structure of APRIL bound to TACI | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
1XU2
 
 | | The crystal structure of APRIL bound to BCMA | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
3DVG
 
 | |
3DVN
 
 | |
2RAW
 
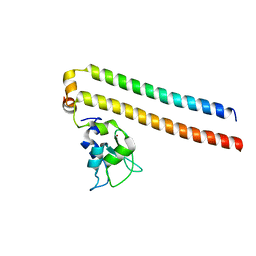 | | Crystal structure of the Borealin-Survivin complex | | Descriptor: | Baculoviral IAP repeat-containing protein 5, Borealin, ZINC ION | | Authors: | Hymowitz, S.G. | | Deposit date: | 2007-09-17 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The mitotic regulator Survivin binds as a monomer to its functional interactor Borealin.
J.Biol.Chem., 282, 2007
|
|
2RAX
 
 | |
4G3E
 
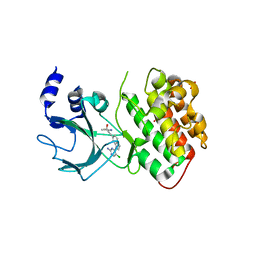 | |
4G3F
 
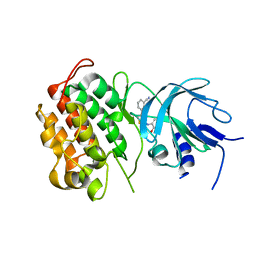 | |
4G3D
 
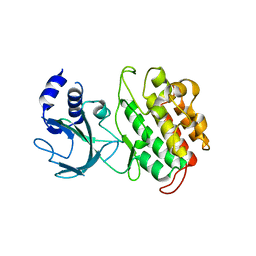 | |
4G3C
 
 | |
3HL5
 
 | | Crystal structure of XIAP BIR3 with CS3 | | Descriptor: | (3S)-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-3-methyl-N-(2-pyrimidin-2-ylphenyl)-L-prolinamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Hymowitz, S.G. | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism of c-IAP and XIAP proteins is required for efficient induction of cell death by small-molecule IAP antagonists.
Acs Chem.Biol., 4, 2009
|
|
3HO4
 
 | |
3HO5
 
 | |
3K48
 
 | | Crystal structure of APRIL bound to a peptide | | Descriptor: | Tumor necrosis factor ligand superfamily member 13, peptide | | Authors: | Hymowitz, S.G. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Multiple novel classes of APRIL-specific receptor-blocking peptides isolated by phage display.
J.Mol.Biol., 396, 2010
|
|
3L95
 
 | |
3MXV
 
 | |
3MXW
 
 | |
2HEY
 
 | | Crystal structure of murine OX40L bound to human OX40 | | Descriptor: | SULFATE ION, Tumor necrosis factor ligand superfamily member 4, Tumor necrosis factor receptor superfamily member 4 | | Authors: | Hymowitz, S.G. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Costimulatory OX40-OX40L Complex.
Structure, 14, 2006
|
|
2H9G
 
 | |
