8I7Q
 
 | |
6VRG
 
 | | Structure of HIV-1 integrase with native amino-terminal sequence | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Eilers, G, Gupta, K, Allen, A, Zhou, J, Hwang, Y, Cory, M, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2020-02-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influence of the amino-terminal sequence on the structure and function of HIV integrase.
Retrovirology, 17, 2020
|
|
1HYV
 
 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH TETRAPHENYL ARSONIUM | | Descriptor: | CHLORIDE ION, INTEGRASE, SULFATE ION, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HYZ
 
 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH A DERIVATIVE OF TETRAPHENYL ARSONIUM. | | Descriptor: | (3,4-DIHYDROXY-PHENYL)-TRIPHENYL-ARSONIUM, CHLORIDE ION, INTEGRASE, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5E6F
 
 | | Canarypox virus resolvase | | Descriptor: | CNPV261 Holliday junction resolvase protein, D(-)-TARTARIC ACID, MAGNESIUM ION | | Authors: | Li, H, Hwang, Y, Perry, K, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Metal Binding Properties of a Poxvirus Resolvase.
J.Biol.Chem., 291, 2016
|
|
2H7F
 
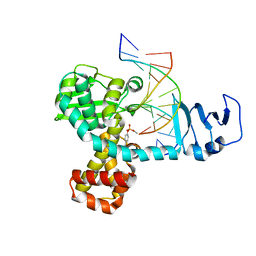 | | Structure of variola topoisomerase covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*T)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
2H7G
 
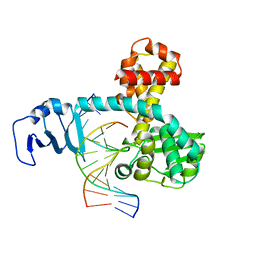 | | Structure of variola topoisomerase non-covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*TP*A)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
5HOT
 
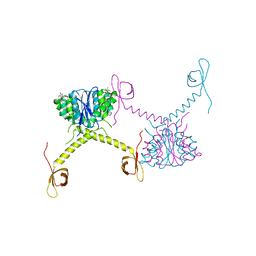 | | Structural Basis for Inhibitor-Induced Aggregation of HIV-1 Integrase | | Descriptor: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, Integrase | | Authors: | Gupta, K, Turkki, V, Sherrill-Mix, S, Hwang, Y, Eilers, G, Taylor, L, McDanal, C, Wang, P, Temelkoff, D, Nolte, R, Velthuisen, E, Jeffrey, J, Van Duyne, G.D, Bushman, F.D. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
3BIY
 
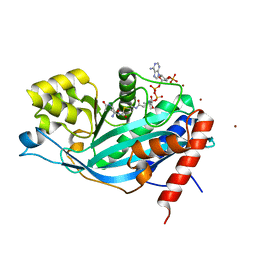 | | Crystal structure of p300 histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | BROMIDE ION, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Liu, X, Wang, L, Zhao, K, Thompson, P.R, Hwang, Y, Marmorstein, R, Cole, P.A. | | Deposit date: | 2007-12-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of protein acetylation by the p300/CBP transcriptional coactivator
Nature, 451, 2008
|
|
3IGC
 
 | | Smallpox virus topoisomerase-DNA transition state | | Descriptor: | 5'-D(*AP*TP*TP*CP*C)-3', 5'-D(*CP*GP*GP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*GP*TP*GP*TP*CP*GP*CP*CP*CP*TP*T)-3', ... | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2009-07-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic.
Structure, 18, 2010
|
|
4OJR
 
 | | Crystal Structure of the HIV-1 Integrase catalytic domain with GSK1264 | | Descriptor: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, CACODYLATE ION, HIV-1 Integrase, ... | | Authors: | Gupta, K, Brady, T, Dyer, B, Hwang, Y, Male, F, Nolte, R.T, Wang, L, Velthuisen, E, Jeffrey, J, Van Duyne, G, Bushman, F.D. | | Deposit date: | 2014-01-21 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Allosteric Inhibition of Human Immunodeficiency Virus Integrase: LATE BLOCK DURING VIRAL REPLICATION AND ABNORMAL MULTIMERIZATION INVOLVING SPECIFIC PROTEIN DOMAINS.
J.Biol.Chem., 289, 2014
|
|
7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
8CT5
 
 | |
8CT7
 
 | | Catalytic Core Domain of HIV-1 Integrase (F185K) bound with BI-224436 | | Descriptor: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
8CTA
 
 | | Minimal 2:2 Ternary Complex between BI-224436 bound HIV-1 Integrase Catalytic Core Domain Dimer and Carboxy Terminal Domains | | Descriptor: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase | | Authors: | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
7KMG
 
 | | LY-CoV555 neutralizing antibody against SARS-CoV-2 | | Descriptor: | GLYCEROL, LY-CoV555 Fab heavy chain, LY-CoV555 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
7KMH
 
 | | LY-CoV488 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LY-CoV488 Fab heavy chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Boyles, J.S, Dickinson, C.D, Coleman, K.A. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
7KMI
 
 | | LY-CoV481 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LY-CoV481 Fab heavy chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
7L3N
 
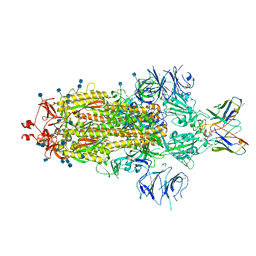 | | SARS-CoV 2 Spike Protein bound to LY-CoV555 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV555 Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection.
Biorxiv, 2020
|
|
5HRR
 
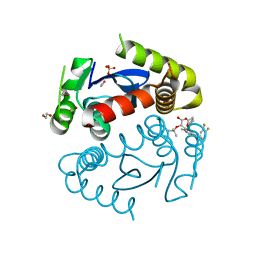 | | HIV Integrase Catalytic Domain containing F185K + A124N + T125S mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5HRP
 
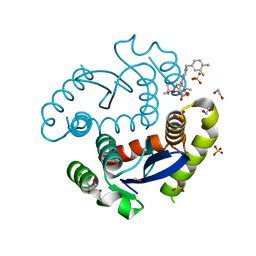 | | HIV Integrase Catalytic Domain containing F185K + A124T mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5HRS
 
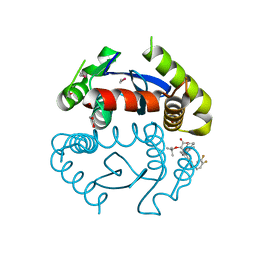 | | HIV Integrase Catalytic Domain containing F185K + A124N + T125A mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5HRN
 
 | | HIV Integrase Catalytic Domain containing F185K mutation complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-23 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
7YGK
 
 | |
6L2U
 
 | | Soluble methane monooxygenase reductase FAD-binding domain from Methylosinus sporium. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methane monooxygenase | | Authors: | Park, J.H, Ha, S.C, Rao, Z, Yoo, H, Yoon, C, Kim, S.Y, Kim, D.S, Lee, S.J. | | Deposit date: | 2019-10-07 | | Release date: | 2021-03-03 | | Last modified: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of the electron transfer environment in the MMOR FAD-binding domain from Methylosinus sporium 5.
Dalton Trans, 50, 2021
|
|
