6BWZ
 
 | | SYSGYS from low-complexity domain of FUS, residues 37-42 | | Descriptor: | SYSGYS peptide from low-complexity domain of FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Tamir, G, Eisenberg, D.S. | | Deposit date: | 2017-12-15 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BZP
 
 | | STGGYG from low-complexity domain of FUS, residues 77-82 | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, RNA-binding protein FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BXV
 
 | | SYSSYGQS from low-complexity domain of FUS, residues 54-61 | | Descriptor: | FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-19 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BZM
 
 | | GFGNFGTS from low-complexity/FG repeat domain of Nup98, residues 116-123 | | Descriptor: | Nuclear pore complex protein Nup98-Nup96 | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Chong, L, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-24 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BXX
 
 | | GYNGFG from low-complexity domain of hnRNPA1, residues 243-248 | | Descriptor: | hnRNPA1 | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-19 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
8TH5
 
 | |
8TH1
 
 | |
8TH7
 
 | |
8TH6
 
 | |
7N8R
 
 | | FGTGFG segment from the Nucleoporin p54, residues 63-68 | | Descriptor: | FGTGFG segment from the Nucleoporin p54, residues 63-68, trifluoroacetic acid | | Authors: | Hughes, M.P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Extended beta-Strands Contribute to Reversible Amyloid Formation.
Acs Nano, 16, 2022
|
|
6WQK
 
 | | hnRNPA2 Low complexity domain (LCD) determined by cryoEM | | Descriptor: | MCherry fluorescent protein,Heterogeneous nuclear ribonucleoproteins A2/B1 chimera | | Authors: | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
6WPQ
 
 | | GNYNVF from hnRNPA2-low complexity domain segment, residues 286-291, D290V variant | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A2 | | Authors: | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
8V1L
 
 | |
7K3Y
 
 | | GGYAGAS segment 52-58 from Keratin-8 | | Descriptor: | GGYAGAS segment 52-58 from the low complexity domain of Keratin-8 | | Authors: | Murray, K.A, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2020-09-14 | | Release date: | 2021-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identifying amyloid-related diseases by mapping mutations in low-complexity protein domains to pathologies.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7K3C
 
 | | SGMGGIT segment 58-64 from Keratin-8 | | Descriptor: | ETHANOL, SGMGGIT segment 58-64 from the low complexity domain of Keratin-8, TRIETHYLENE GLYCOL | | Authors: | Murray, K.A, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2020-09-11 | | Release date: | 2021-11-10 | | Last modified: | 2022-07-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identifying amyloid-related diseases by mapping mutations in low-complexity protein domains to pathologies.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7K3X
 
 | |
6B79
 
 | |
8DUW
 
 | | HnRNPA2 D290V LCD PM2 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
8DU2
 
 | | HnRNPA2 D290V LCD PM1 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
8EC7
 
 | | HnRNPA2 D290V LCD PM3 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
5WMJ
 
 | |
5WOR
 
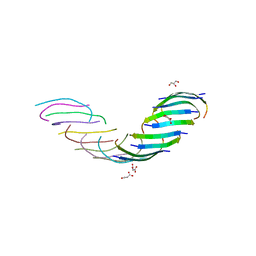 | |
6CFH
 
 | |
6CB9
 
 | | Segment AALQSS from the low complexity domain of TDP-43, residues 328-333 | | Descriptor: | AALQSS | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-02 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CEW
 
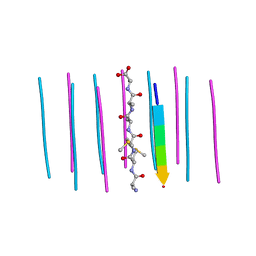 | | Segment AMMAAA from the low complexity domain of TDP-43, residues 321-326 | | Descriptor: | AMMAAA | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
