4OQZ
 
 | | Streptomyces aurantiacus imine reductase | | Descriptor: | Putative oxidoreductase YfjR | | Authors: | Schneider, L.K, Huber, T, Gerhardt, S, Muller, M, Einsle, O. | | Deposit date: | 2014-02-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Direct Reductive Amination of Ketones: Structure and Activity of S-Selective Imine Reductases from Streptomyces.
CHEMCATCHEM, 2014
|
|
4OQY
 
 | | Streptomyces sp. GF3546 imine reductase | | Descriptor: | (S)-imine reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schneider, L.K, Huber, T, Gerhardt, S, Muller, M, Einsle, O. | | Deposit date: | 2014-02-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Direct Reductive Amination of Ketones: Structure and Activity of S-Selective Imine Reductases from Streptomyces.
CHEMCATCHEM, 2014
|
|
6O6I
 
 | |
7R6O
 
 | | Pyrrolysyl-tRNA synthetase from methanogenic archaeon ISO4-G1 (G1PylRS) | | Descriptor: | 1,2-ETHANEDIOL, Pyrrolysyl-tRNA synthetase PylS, SULFATE ION | | Authors: | Frkic, R.L, Huber, T, Jackson, C.J. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genetic Encoding of Cyanopyridylalanine for In-Cell Protein Macrocyclization by the Nitrile-Aminothiol Click Reaction.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
2M66
 
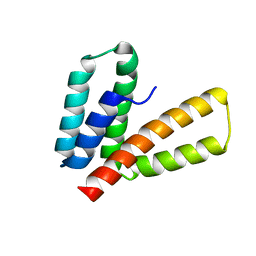 | | Endoplasmic reticulum protein 29 (ERp29) C-terminal domain: 3D Protein Fold Determination from Backbone Amide Pseudocontact Shifts Generated by Lanthanide Tags at Multiple Sites | | Descriptor: | Endoplasmic reticulum resident protein 29 | | Authors: | Yagi, H, Pilla, K, Maleckis, A, Graham, B, Huber, T, Otting, G. | | Deposit date: | 2013-03-26 | | Release date: | 2013-07-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional protein fold determination from backbone amide pseudocontact shifts generated by lanthanide tags at multiple sites
Structure, 21, 2013
|
|
1WNH
 
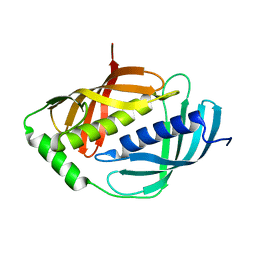 | | Crystal structure of mouse Latexin (tissue carboxypeptidase inhibitor) | | Descriptor: | Latexin | | Authors: | Aagaard, A, Listwan, P, Cowieson, N, Huber, T, Ravasi, T, Wells, C.A, Flanagan, J.U, Hume, D.A, Kobe, B, Martin, J.L. | | Deposit date: | 2004-08-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An Inflammatory Role for the Mammalian Carboxypeptidase Inhibitor Latexin: Relationship to Cystatins and the Tumor Suppressor TIG1
Structure, 13, 2005
|
|
2V7N
 
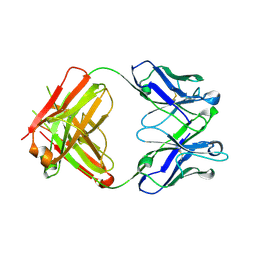 | | Unusual twinning in crystals of the CitS binding antibody Fab fragment f3p4 | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Frey, D, Huber, T, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2007-07-31 | | Release date: | 2008-06-17 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the Recombinant Antibody Fab Fragment F3P4.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2V1O
 
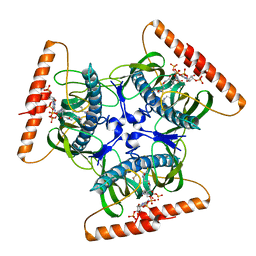 | | Crystal structure of N-terminal domain of acyl-CoA thioesterase 7 | | Descriptor: | COENZYME A, CYTOSOLIC ACYL COENZYME A THIOESTER HYDROLASE | | Authors: | Forwood, J.K, Thakur, A.S, Guncar, G, Marfori, M, Mouradov, D, Meng, W.N, Robinson, J, Huber, T, Kellie, S, Martin, J.L, Hume, D.A, Kobe, B. | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Recruitment of Tandem Hotdog Domains in Acyl-Coa Thioesterase 7 and its Role in Inflammation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
7T2Q
 
 | | PEGylated Calmodulin-1 (K148U) | | Descriptor: | CALCIUM ION, Calmodulin-1, MAGNESIUM ION, ... | | Authors: | Mackay, J.P, Payne, R.J, Patel, K, Dowman, L.J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Site-selective photocatalytic functionalization of peptides and proteins at selenocysteine.
Nat Commun, 13, 2022
|
|
7Z3W
 
 | | Crystal structure of the AAL160 Fab | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AAL160 Fab heavy-chain, AAL160 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2022-03-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | "Redirecting an anti-IL-1 beta antibody to bind a new, unrelated and computationally predicted epitope on hIL-17A".
Commun Biol, 6, 2023
|
|
7Z2M
 
 | |
7Z4T
 
 | | AAL160 FAB IN COMPLEX WITH HUMAN INTERLEUKIN-1 BETA | | Descriptor: | AAL160 Fab heavy-chain, AAL160 light-chain, Interleukin-1 beta | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2022-03-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | "Redirecting an anti-IL-1 beta antibody to bind a new, unrelated and computationally predicted epitope on hIL-17A".
Commun Biol, 6, 2023
|
|
4W1P
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 5.54 MGy TEMP 150K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-14 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4W1Q
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 7.39 MGy TEMP 150K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-14 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4W1S
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 11.09 MGy TEMP 150K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | jackson, C.j, carr, p.d, weik, m, huber, t, meirelles, t, correy, g. | | Deposit date: | 2014-08-14 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
7RFD
 
 | |
7N3J
 
 | |
6ZTB
 
 | | Crystal Structure of human P-Cadherin EC1_EC2 | | Descriptor: | CALCIUM ION, Cadherin-3, SODIUM ION | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTF
 
 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 | | Descriptor: | CQY684 Fab heavy-chain, CQY684 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTR
 
 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 in complex with human P-Cadherin(108-324) | | Descriptor: | CALCIUM ION, CQY684 Fab heavy-chain, CQY684 Fab light-chain, ... | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
4UBO
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 3.70 MGy TEMP 150K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4UBJ
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 5.55 MGy at 100K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4UBK
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 7.40 MGy at 100K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4UBM
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 11.11 MGy at 100K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4UBI
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 3.70 MGy at 100K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
