3WRE
 
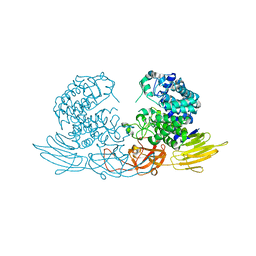 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRG
 
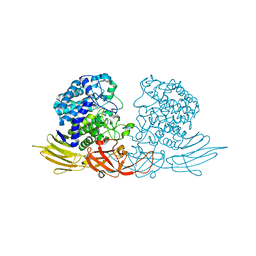 | | The complex structure of HypBA1 with L-arabinose | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRF
 
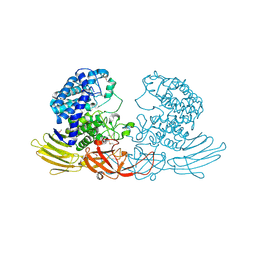 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
7BSL
 
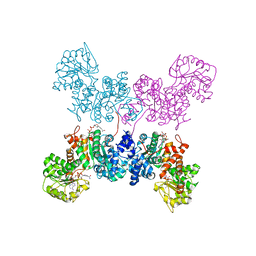 | | Crystal Structure of human ME2 R67A mutant | | Descriptor: | NAD-dependent malic enzyme, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, W.L, Tai, S.C, Hung, H.C, Ho, M.C. | | Deposit date: | 2020-03-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Single nucleotide variants lead to dysregulation of the human mitochondrial NAD(P) + -dependent malic enzyme.
Iscience, 24, 2021
|
|
7BSK
 
 | | Crystal structure of human ME2 R67Q mutant | | Descriptor: | NAD-dependent malic enzyme, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, W.L, Tai, S.C, Hung, H.C, Ho, M.C. | | Deposit date: | 2020-03-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Single nucleotide variants lead to dysregulation of the human mitochondrial NAD(P) + -dependent malic enzyme.
Iscience, 24, 2021
|
|
7BSJ
 
 | | Crystal structure of human ME2 R484W | | Descriptor: | FUMARIC ACID, MAGNESIUM ION, NAD-dependent malic enzyme, ... | | Authors: | Chen, W.L, Tai, S.C, Hung, H.C, Ho, M.C. | | Deposit date: | 2020-03-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Single nucleotide variants lead to dysregulation of the human mitochondrial NAD(P) + -dependent malic enzyme.
Iscience, 24, 2021
|
|
