2HKY
 
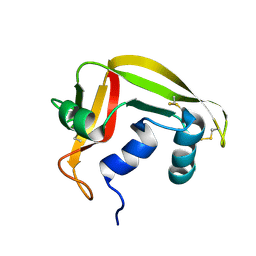 | | NMR solution structure of human RNase 7 | | Descriptor: | Ribonuclease 7 | | Authors: | Huang, Y.-C, Chen, C, Lou, Y.-C. | | Deposit date: | 2006-07-06 | | Release date: | 2006-12-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity.
J.Biol.Chem., 282, 2007
|
|
3GYX
 
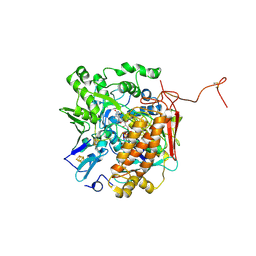 | | Crystal structure of adenylylsulfate reductase from Desulfovibrio gigas | | Descriptor: | Adenylylsulfate Reductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER | | Authors: | Chiang, Y.-L, Hsieh, Y.-C, Liu, E.-H, Liu, M.-Y, Chen, C.-J. | | Deposit date: | 2009-04-06 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Adenylylsulfate reductase from Desulfovibrio gigas suggests a potential self-regulation mechanism involving the C terminus of the beta-subunit
J.Bacteriol., 191, 2009
|
|
3N13
 
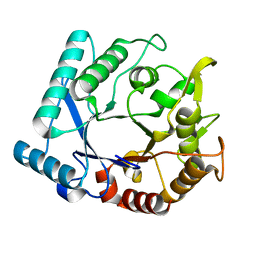 | | Crystal stricture of D143A chitinase in complex with NAG from Bacillus cereus NCTU2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Hsieh, Y.-C, Wu, Y.-J, Wu, W.-G, Li, Y.-K, Chen, C.-J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of bacillus cereus NCTU2 chitinase complexes with chitooligomers reveal novel substrate binding for catalysis: a chitinase without chitin-binding and insertion domains
J.Biol.Chem., 285, 2010
|
|
3N18
 
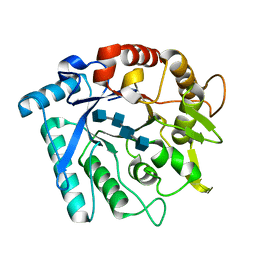 | | Crystal stricture of E145G/Y227F chitinase in complex with NAG from Bacillus cereus NCTU2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Hsieh, Y.-C, Wu, Y.-J, Wu, W.-G, Li, Y.-K, Chen, C.-J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of bacillus cereus NCTU2 chitinase complexes with chitooligomers reveal novel substrate binding for catalysis: a chitinase without chitin-binding and insertion domains
J.Biol.Chem., 285, 2010
|
|
3N17
 
 | | Crystal stricture of E145Q/Y227F chitinase in complex with NAG from Bacillus cereus NCTU2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Hsieh, Y.-C, Wu, Y.-J, Wu, W.-G, Li, Y.-K, Chen, C.-J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of bacillus cereus NCTU2 chitinase complexes with chitooligomers reveal novel substrate binding for catalysis: a chitinase without chitin-binding and insertion domains
J.Biol.Chem., 285, 2010
|
|
3N11
 
 | |
3N15
 
 | | Crystal stricture of E145Q chitinase in complex with NAG from Bacillus cereus NCTU2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Hsieh, Y.-C, Wu, Y.-J, Wu, W.-G, Li, Y.-K, Chen, C.-J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of bacillus cereus NCTU2 chitinase complexes with chitooligomers reveal novel substrate binding for catalysis: a chitinase without chitin-binding and insertion domains
J.Biol.Chem., 285, 2010
|
|
3N12
 
 | | Crystal stricture of chitinase in complex with zinc atoms from Bacillus cereus NCTU2 | | Descriptor: | ACETIC ACID, Chitinase A, ZINC ION | | Authors: | Hsieh, Y.-C, Wu, Y.-J, Wu, W.-G, Li, Y.-K, Chen, C.-J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of bacillus cereus NCTU2 chitinase complexes with chitooligomers reveal novel substrate binding for catalysis: a chitinase without chitin-binding and insertion domains
J.Biol.Chem., 285, 2010
|
|
3N1A
 
 | | Crystal stricture of E145G/Y227F chitinase in complex with cyclo-(L-His-L-Pro) from Bacillus cereus NCTU2 | | Descriptor: | CYCLO-(L-HISTIDINE-L-PROLINE) INHIBITOR, Chitinase A | | Authors: | Hsieh, Y.-C, Wu, Y.-J, Wu, W.-G, Li, Y.-K, Chen, C.-J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of bacillus cereus NCTU2 chitinase complexes with chitooligomers reveal novel substrate binding for catalysis: a chitinase without chitin-binding and insertion domains
J.Biol.Chem., 285, 2010
|
|
2DSX
 
 | | Crystal structure of rubredoxin from Desulfovibrio gigas to ultra-high 0.68 A resolution | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Chen, C.-J, Lin, Y.-H, Huang, Y.-C, Liu, M.-Y. | | Deposit date: | 2006-07-07 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.68 Å) | | Cite: | Crystal structure of rubredoxin from Desulfovibrio gigas to ultra-high 0.68A resolution
Biochem.Biophys.Res.Commun., 349, 2006
|
|
7XGS
 
 | | Short vegetative phase protein | | Descriptor: | 1,2-ETHANEDIOL, MADS-box protein SVP | | Authors: | Liao, Y.-T, Wang, H.-C, Ko, T.-P. | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The PHYL1 effector of peanut witches broom (PnWB) phytoplasma alters the miR156/157-mediated squamosa promoter binding protein like regulation and gibberellic acid generation to promote anthocyanin biosynthesis
To Be Published
|
|
