1D2G
 
 | | CRYSTAL STRUCTURE OF R175K MUTANT GLYCINE N-METHYLTRANSFERASE FROM RAT LIVER | | Descriptor: | GLYCINE N-METHYLTRANSFERASE | | Authors: | Huang, Y, Komoto, J, Takusagawa, F, Konishi, K, Takata, Y. | | Deposit date: | 1999-10-08 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1D2H
 
 | | CRYSTAL STRUCTURE OF R175K MUTANT GLYCINE N-METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLHOMOCYSTEINE | | Descriptor: | GLYCINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Huang, Y, Komoto, J, Takusagawa, F, Konishi, K, Takata, Y. | | Deposit date: | 1999-10-11 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1D2C
 
 | | METHYLTRANSFERASE | | Descriptor: | PROTEIN (GLYCINE N-METHYLTRANSFERASE) | | Authors: | Huang, Y, Takusagawa, F. | | Deposit date: | 1999-09-23 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1OMY
 
 | | Crystal Structure of a Recombinant alpha-insect Toxin BmKaIT1 from the scorpion Buthus martensii Karsch | | Descriptor: | ACETIC ACID, Alpha-neurotoxin TX12, CHLORIDE ION | | Authors: | Huang, Y, Huang, Q, Chen, H, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary crystallographic study of rBmKalphaIT1, a recombinant alpha-insect toxin from the scorpion Buthus martensii Karsch.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1I4O
 
 | | CRYSTAL STRUCTURE OF THE XIAP/CASPASE-7 COMPLEX | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, CASPASE-7 | | Authors: | Huang, Y, Park, Y.C, Rich, R.L, Segal, D, Myszka, D.G, Wu, H. | | Deposit date: | 2001-02-22 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain.
Cell(Cambridge,Mass.), 104, 2001
|
|
3HTX
 
 | | Crystal structure of small RNA methyltransferase HEN1 | | Descriptor: | 5'-R(*GP*AP*UP*UP*UP*CP*UP*CP*UP*CP*UP*GP*CP*AP*AP*GP*CP*GP*AP*AP*AP*G)-3', 5'-R(P*UP*UP*CP*GP*CP*UP*UP*GP*CP*AP*GP*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*C)-3', HEN1, ... | | Authors: | Huang, Y, Ji, L.-J, Huang, Q.-C, Vassylyev, D.G, Chen, X.-M, Ma, J.-B. | | Deposit date: | 2009-06-12 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into mechanisms of the small RNA methyltransferase HEN1.
Nature, 461, 2009
|
|
1PW4
 
 | | Crystal Structure of the Glycerol-3-Phosphate Transporter from E.Coli | | Descriptor: | Glycerol-3-phosphate transporter | | Authors: | Huang, Y, Lemieux, M.J, Song, J, Auer, M, Wang, D.N. | | Deposit date: | 2003-06-30 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and Mechanism of the Glycerol-3-Phosphate Transporter from Escherichia Coli
Science, 301, 2003
|
|
3FIP
 
 | | Crystal structure of Usher PapC translocation pore | | Descriptor: | Outer membrane usher protein papC | | Authors: | Huang, Y, Deisenhofer, J. | | Deposit date: | 2008-12-12 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.154 Å) | | Cite: | Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2FMM
 
 | | Crystal Structure of EMSY-HP1 complex | | Descriptor: | Chromobox protein homolog 1, Protein EMSY, SULFATE ION | | Authors: | Huang, Y. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the HP1-EMSY complex reveals an unusual mode of HP1 binding.
Structure, 14, 2006
|
|
3MJ0
 
 | |
7SZ7
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to TGF-alpha. "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor receptor, Transforming growth factor alpha | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ0
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ1
 
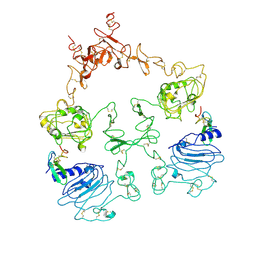 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ5
 
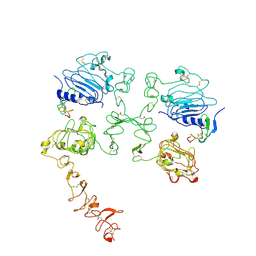 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to TGF-alpha "tips-separated" conformation | | Descriptor: | Epidermal growth factor receptor, Transforming growth factor alpha | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYE
 
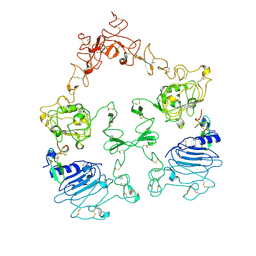 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYD
 
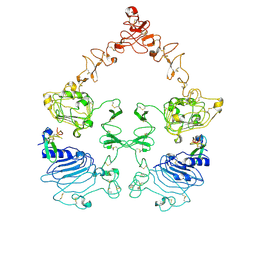 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7UGX
 
 | | Asp-bound GltPh RSMR mutant in IFS-B1 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19 F NMR and Cryo-EM.
J.Am.Chem.Soc., 145, 2023
|
|
7UG0
 
 | | TBOA-bound GltPh RSMR mutant in IFS state | | Descriptor: | (3S)-3-(BENZYLOXY)-L-ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UGD
 
 | | Asp-bound GltPh RSMR mutant in IFS-A1 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UH3
 
 | | Asp-bound GltPh RSMR mutant in iOFS state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UGJ
 
 | | TBOA-bound GltPh RSMR mutant in OFS state | | Descriptor: | (3S)-3-(BENZYLOXY)-L-ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
7UGV
 
 | | Asp-bound GltPh RSMR mutant in IFS-A2 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19 F NMR and Cryo-EM.
J.Am.Chem.Soc., 145, 2023
|
|
7UH6
 
 | | Asp-bound GltPh RSMR mutant in IFS-B2 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
2GF7
 
 | | Double tudor domain structure | | Descriptor: | Jumonji domain-containing protein 2A, SULFATE ION | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
8GTC
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C baseplate-tail complex | | Descriptor: | Distal tail protein, Hub protein, Major tail protein, ... | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
