2AXR
 
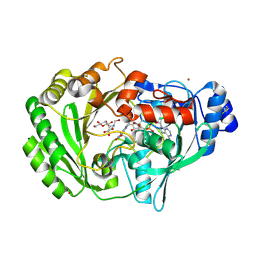 | | Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8alpha-N1-histidyl FAD | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Huang, C.-H, Lai, W.-L, Vasella, A, Tsai, Y.-C, Liaw, S.-H. | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of Glucooligosaccharide Oxidase from Acremonium strictum: A NOVEL FLAVINYLATION OF 6-S-CYSTEINYL, 8{alpha}-N1-HISTIDYL FAD
J.Biol.Chem., 280, 2005
|
|
3HSU
 
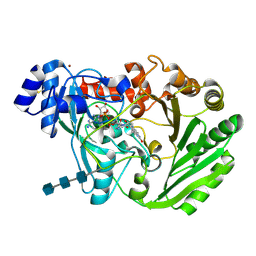 | | Functional roles of the 6-s-cysteinyl, 8 alpha-N1-histidyl FAD in Glucooligosaccharide Oxidase from Acremonium strictum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Liaw, S.-H, Huang, C.-H. | | Deposit date: | 2009-06-10 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Functional roles of the 6-S-cysteinyl, 8alpha-N1-histidyl FAD in glucooligosaccharide oxidase from Acremonium strictum
J.Biol.Chem., 283, 2008
|
|
3VKA
 
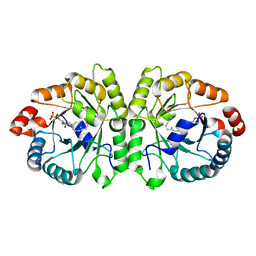 | | Crystal structure of MoeO5 soaked for 3 hours in FsPP | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5 | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VKB
 
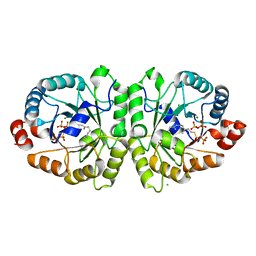 | | Crystal structure of MoeO5 soaked with FsPP overnight | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MoeO5, ... | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VKD
 
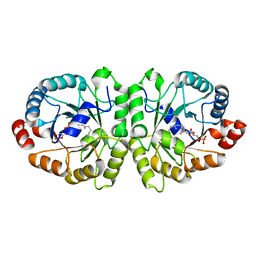 | | Crystal structure of MoeO5 soaked with 3-phosphoglycerate | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5, ... | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VK5
 
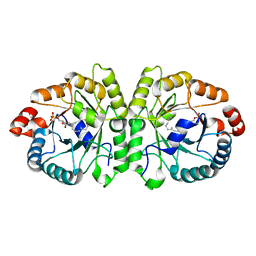 | | Crystal structure of MoeO5 in complex with its product FPG | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5 | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-08 | | Release date: | 2012-05-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3W28
 
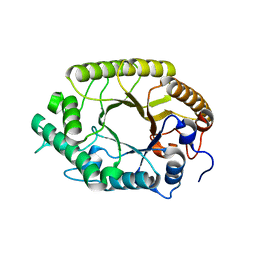 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W29
 
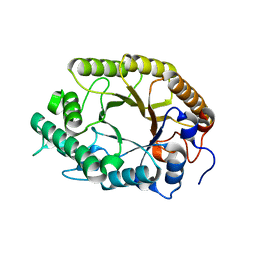 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3VKC
 
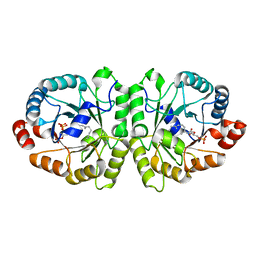 | | Crystal structure of MoeO5 soaked with pyrophosphate | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5, ... | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3W27
 
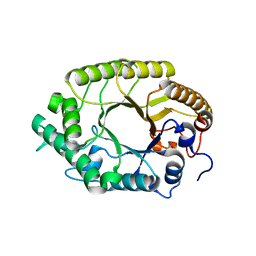 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W25
 
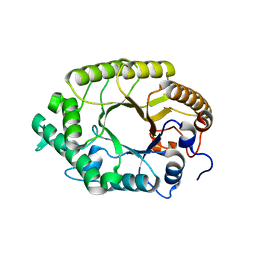 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W26
 
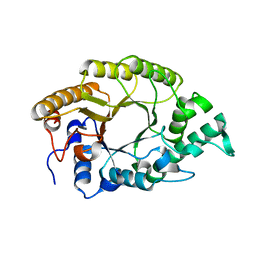 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W24
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 | | Descriptor: | Glycoside hydrolase family 10 | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
1ZR6
 
 | | The crystal structure of an Acremonium strictum glucooligosaccharide oxidase reveals a novel flavinylation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ZINC ION, ... | | Authors: | Huang, C.-H, Lai, W.-L, Lee, M.-H, Tsai, Y.-C, Liaw, S.-H. | | Deposit date: | 2005-05-19 | | Release date: | 2005-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8alpha-N1-histidyl FAD
J.Biol.Chem., 280, 2005
|
|
3AYS
 
 | | GH5 endoglucanase from a ruminal fungus in complex with cellotriose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Tseng, C.-W, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | Deposit date: | 2011-05-16 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate binding of a GH5 endoglucanase from the ruminal fungus Piromyces rhizinflata.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3AYR
 
 | | GH5 endoglucanase EglA from a ruminal fungus | | Descriptor: | Endoglucanase | | Authors: | Tseng, C.-W, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | Deposit date: | 2011-05-16 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate binding of a GH5 endoglucanase from the ruminal fungus Piromyces rhizinflata.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3AMH
 
 | | crystal structure of cellulase 12A from Thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMP
 
 | | E134C-Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMQ
 
 | | E134C-Cellobiose co-crystal of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMM
 
 | | Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMN
 
 | | E134C-Cellobiose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3WGH
 
 | | Crystal structure of RSP in complex with beta-NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CACODYLATE ION, Redox-sensing transcriptional repressor rex, ... | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
3WGI
 
 | | Crystal structure of RSP in complex with beta-NAD+ and operator DNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*TP*GP*TP*TP*AP*AP*TP*CP*GP*AP*TP*TP*AP*AP*CP*AP*AP*TP*C)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), Redox-sensing transcriptional repressor rex | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
3WG9
 
 | | Crystal structure of RSP, a Rex-family repressor | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
