6JQZ
 
 | | ZHD/H242A complex with ZEN | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, GLYCEROL, Zearalenone hydrolase | | Authors: | Hu, X.J. | | Deposit date: | 2019-04-02 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of ZHD complex
To Be Published
|
|
6JRA
 
 | | ZHD/W183F complex with hydrolyzed aZOL | | Descriptor: | 2-[(~{E},6~{R},10~{S})-6,10-bis(oxidanyl)undec-1-enyl]-4,6-bis(oxidanyl)benzoic acid, GLYCEROL, POTASSIUM ION, ... | | Authors: | Hu, X.J. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ZHD complex
To Be Published
|
|
6JRC
 
 | | ZHD complex with hydrolyzed alpha-ZOL | | Descriptor: | 2-[(~{E},6~{R},10~{S})-6,10-bis(oxidanyl)undec-1-enyl]-4,6-bis(oxidanyl)benzoic acid, GLYCEROL, POTASSIUM ION, ... | | Authors: | Hu, X.J. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of ZHD complex
To Be Published
|
|
6JRB
 
 | | ZHD/W183F complex with bZOL | | Descriptor: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, GLYCEROL, Zearalenone hydrolase | | Authors: | Hu, X.J. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ZHD complex
To Be Published
|
|
6JRD
 
 | | ZHD complex with hydrolyzed beta-ZOL | | Descriptor: | 2-[(~{E},6~{S},10~{S})-6,10-bis(oxidanyl)undec-1-enyl]-4,6-bis(oxidanyl)benzoic acid, GLYCEROL, POTASSIUM ION, ... | | Authors: | Hu, X.J. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of ZHD complex
To Be Published
|
|
6JR2
 
 | | ZHD/H242A complex with aZOL | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, GLYCEROL, POTASSIUM ION, ... | | Authors: | Hu, X.J. | | Deposit date: | 2019-04-02 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of ZHD complex
To Be Published
|
|
6JR5
 
 | | ZHD/H242A complex with bZOL | | Descriptor: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, GLYCEROL, Zearalenone hydrolase | | Authors: | Hu, X.J. | | Deposit date: | 2019-04-02 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of ZHD complex
To Be Published
|
|
6JR9
 
 | | ZHD/W183F complex with ZEN | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, GLYCEROL, POTASSIUM ION, ... | | Authors: | Hu, X.J. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of ZHD complex
To Be Published
|
|
5JVA
 
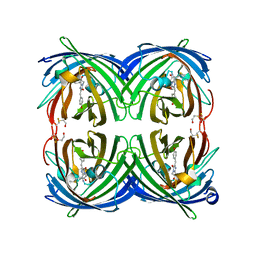 | | 1.95 angstrom crystal structure of TAGRFP-T | | Descriptor: | BETA-MERCAPTOETHANOL, SULFATE ION, TagRFP-T | | Authors: | Hu, X.J. | | Deposit date: | 2016-05-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of red fluorescent protein TagRFP-T reveals the mechanism of its superior photostability.
Biochem. Biophys. Res. Commun., 477, 2016
|
|
5XMW
 
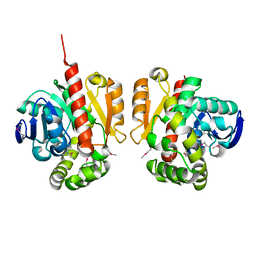 | | Selenomethionine-derivated ZHD | | Descriptor: | Zearalenone lactonase | | Authors: | Hu, X.J. | | Deposit date: | 2017-05-16 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4PXG
 
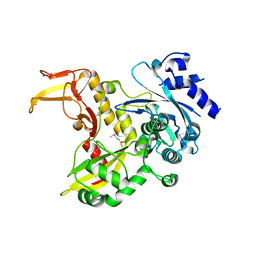 | |
5Z87
 
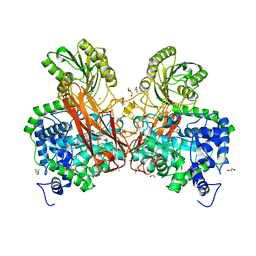 | | Structural of a novel b-glucosidase EmGH1 at 2.3 angstrom from Erythrobacter marinus | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BENZAMIDINE, ... | | Authors: | Li, J.X, Hu, X.J, Zhao, Y, Li, L. | | Deposit date: | 2018-01-31 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical analysis of a novel b-glucosidase EmGH1 from Erythrobacter marinus
To Be Published
|
|
5ZA3
 
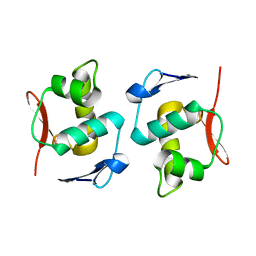 | |
2REU
 
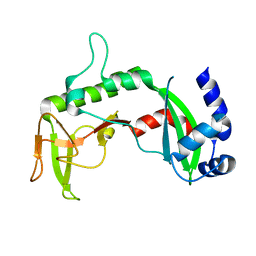 | | Crystal Structure of the C-terminal of Sau3AI fragment | | Descriptor: | MAGNESIUM ION, Type II restriction enzyme Sau3AI | | Authors: | Hu, X, Yu, F, Xu, C, He, J. | | Deposit date: | 2007-09-27 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and function of C-terminal Sau3AI domain
Biochim.Biophys.Acta, 1794, 2009
|
|
5HKE
 
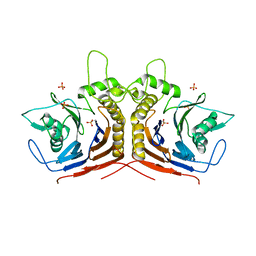 | | bile salt hydrolase from Lactobacillus salivarius | | Descriptor: | Bile salt hydrolase, PHOSPHATE ION | | Authors: | Hu, X.-J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of bile salt hydrolase from Lactobacillus salivarius.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4XO7
 
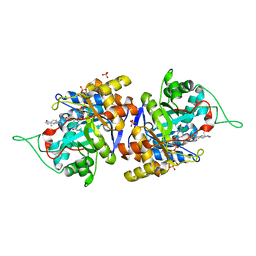 | | Crystal structure of human 3-alpha hydroxysteroid dehydrogenase type 3 in complex with NADP+, 5alpha-androstan-3,17-dione and (3beta, 5alpha)-3-hydroxyandrostan-17-one | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human 3 alpha-hydroxysteroid dehydrogenase type 3: structural clues of 5 alpha-DHT reverse binding and enzyme down-regulation decreasing MCF7 cell growth.
Biochem.J., 473, 2016
|
|
4XO6
 
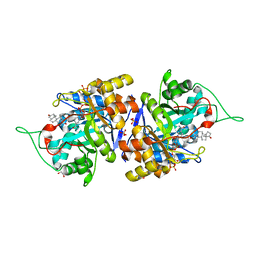 | | Crystal structure of human 3-alpha hydroxysteroid dehydrogenase type 3 in complex with NADP+, 5alpha-androstan-3,17-dione and (3beta, 5alpha)-3-hydroxyandrostan-17-one | | Descriptor: | (3Beta,5alpha)-3-Hydroxyandrostan-17-one, 1,2-ETHANEDIOL, 5ALPHA-ANDROSTAN-3,17-DIONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Human 3 alpha-hydroxysteroid dehydrogenase type 3: structural clues of 5 alpha-DHT reverse binding and enzyme down-regulation decreasing MCF7 cell growth.
Biochem.J., 473, 2016
|
|
5C8Z
 
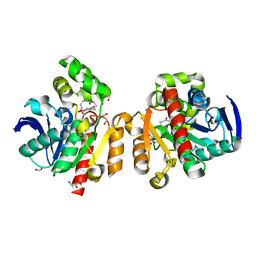 | | ZHD-ZGR complex after ZHD crystal soaking in ZEN for 30min | | Descriptor: | 2,4-dihydroxy-6-[(1E,10S)-10-hydroxy-6-oxoundec-1-en-1-yl]benzoic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X.-J, Qi, Q, Yang, W.-J. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3U1R
 
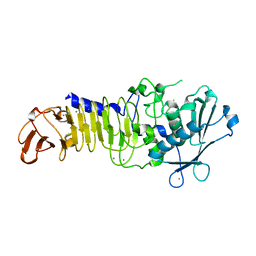 | | Structure Analysis of A New Psychrophilic Marine Protease | | Descriptor: | Alkaline metalloprotease, CALCIUM ION, ZINC ION | | Authors: | Zhang, S.-C, Sun, M, Tang, L, Wang, Q.-H, Hao, J.-H, Han, Y, Hu, X.-J, Zhou, M, Lin, S.-X. | | Deposit date: | 2011-09-30 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure analysis of a new psychrophilic marine protease.
Plos One, 6, 2011
|
|
5Z3K
 
 | |
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
